Gene Page: ACVR2A
Summary ?
| GeneID | 92 |
| Symbol | ACVR2A |
| Synonyms | ACTRII|ACVR2 |
| Description | activin A receptor type 2A |
| Reference | MIM:102581|HGNC:HGNC:173|Ensembl:ENSG00000121989|HPRD:00025|Vega:OTTHUMG00000150603 |
| Gene type | protein-coding |
| Map location | 2q22.3 |
| Pascal p-value | 0.046 |
| Fetal beta | 1.173 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.023 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.02395 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22464182 | 2 | 148602625 | ACVR2A | -0.024 | 0.25 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs5973375 | chrX | 35489060 | ACVR2A | 92 | 0.14 | trans |
Section II. Transcriptome annotation
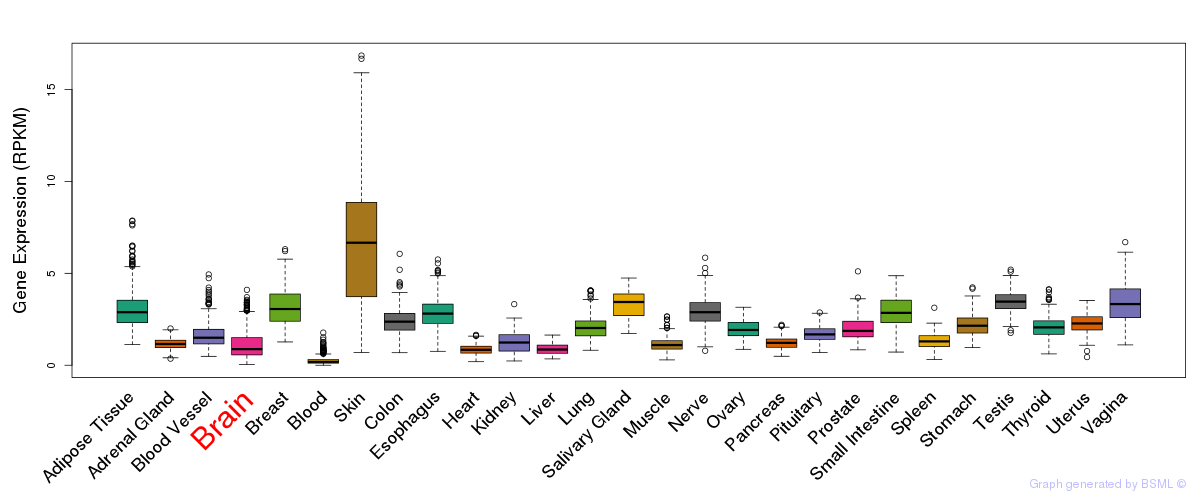
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ABHD14A | 0.96 | 0.95 |
| TEX264 | 0.87 | 0.83 |
| ISOC2 | 0.86 | 0.88 |
| CYC1 | 0.84 | 0.81 |
| FASTK | 0.84 | 0.83 |
| MARCH2 | 0.84 | 0.81 |
| MFSD3 | 0.83 | 0.86 |
| C11orf59 | 0.83 | 0.86 |
| SLC25A11 | 0.83 | 0.78 |
| GPX4 | 0.83 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF5B | -0.60 | -0.70 |
| RBM25 | -0.53 | -0.55 |
| SFRS12 | -0.53 | -0.53 |
| ZNF326 | -0.52 | -0.50 |
| AC005921.3 | -0.49 | -0.69 |
| THOC2 | -0.48 | -0.47 |
| PPIG | -0.47 | -0.45 |
| PRPF38B | -0.47 | -0.47 |
| SFRS18 | -0.46 | -0.58 |
| ESF1 | -0.45 | -0.41 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0019838 | growth factor binding | IEA | - | |
| GO:0017002 | activin receptor activity | IDA | 12665502 | |
| GO:0015026 | coreceptor activity | IDA | 10746731 | |
| GO:0030145 | manganese ion binding | IEA | - | |
| GO:0048185 | activin binding | IPI | 9032295 | |
| GO:0048186 | inhibin beta-A binding | IDA | 7890768 | |
| GO:0043621 | protein self-association | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001702 | gastrulation with mouth forming second | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007368 | determination of left/right symmetry | IEA | - | |
| GO:0007283 | spermatogenesis | IEA | - | |
| GO:0009952 | anterior/posterior pattern formation | IEA | - | |
| GO:0007498 | mesoderm development | IEA | - | |
| GO:0050999 | regulation of nitric-oxide synthase activity | IEA | - | |
| GO:0042713 | sperm ejaculation | IEA | - | |
| GO:0030501 | positive regulation of bone mineralization | IMP | 18436533 | |
| GO:0030509 | BMP signaling pathway | EXP | 15621726 | |
| GO:0030509 | BMP signaling pathway | IDA | 18436533 | |
| GO:0043084 | penile erection | IEA | - | |
| GO:0032927 | positive regulation of activin receptor signaling pathway | IDA | 12665502 | |
| GO:0045648 | positive regulation of erythrocyte differentiation | IDA | 9032295 | |
| GO:0045669 | positive regulation of osteoblast differentiation | IMP | 18436533 | |
| GO:0048706 | embryonic skeletal system development | IEA | - | |
| GO:0060011 | Sertoli cell proliferation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IDA | 14738881 | |
| GO:0005886 | plasma membrane | EXP | 17356069 | |
| GO:0005887 | integral to plasma membrane | TAS | 1314589 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACVR1B | ACTRIB | ACVRLK4 | ALK4 | SKR2 | activin A receptor, type IB | - | HPRD,BioGRID | 8612709 |
| BMP6 | VGR | VGR1 | bone morphogenetic protein 6 | - | HPRD,BioGRID | 10504300 |
| BMP7 | OP-1 | bone morphogenetic protein 7 | - | HPRD,BioGRID | 9748228 |
| ENG | CD105 | END | FLJ41744 | HHT1 | ORW | ORW1 | endoglin | - | HPRD,BioGRID | 9872992 |
| GDF5 | BMP14 | CDMP1 | LAP4 | SYNS2 | growth differentiation factor 5 | - | HPRD,BioGRID | 8702914 |
| GDF9 | - | growth differentiation factor 9 | - | HPRD | 12135884 |
| IGSF1 | IGCD1 | IGDC1 | INHBP | KIAA0364 | MGC75490 | PGSF2 | immunoglobulin superfamily, member 1 | - | HPRD | 11266516 |
| INHA | - | inhibin, alpha | Affinity Capture-Western | BioGRID | 10746731 |
| INHBA | EDF | FRP | inhibin, beta A | Affinity Capture-Western | BioGRID | 9202237 |10746731 |
| INHBA | EDF | FRP | inhibin, beta A | - | HPRD | 11266516 |
| INHBB | MGC157939 | inhibin, beta B | Affinity Capture-Western | BioGRID | 9202237 |
| INHBB | MGC157939 | inhibin, beta B | - | HPRD | 11266516 |
| INHBC | IHBC | inhibin, beta C | - | HPRD | 12651901 |
| MAGI2 | ACVRIP1 | AIP1 | ARIP1 | MAGI-2 | SSCAM | membrane associated guanylate kinase, WW and PDZ domain containing 2 | - | HPRD | 10681527 |
| SYNJ2BP | ARIP2 | FLJ11271 | FLJ41973 | OMP25 | synaptojanin 2 binding protein | - | HPRD,BioGRID | 11882656 |15451561 |
| TGFBR3 | BGCAN | betaglycan | transforming growth factor, beta receptor III | - | HPRD | 10746731 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| PID ALK1 PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NODAL | 18 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY BMP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD DN | 84 | 63 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| CHIARETTI ACUTE LYMPHOBLASTIC LEUKEMIA ZAP70 | 67 | 33 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR DN | 53 | 40 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 UP | 84 | 55 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 11 | 57 | 40 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 9 | 92 | 59 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| MATZUK CENTRAL FOR FEMALE FERTILITY | 29 | 25 | All SZGR 2.0 genes in this pathway |
| MATZUK MALE REPRODUCTION SERTOLI | 28 | 23 | All SZGR 2.0 genes in this pathway |
| BONCI TARGETS OF MIR15A AND MIR16 1 | 91 | 75 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION A | 67 | 52 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY UP | 66 | 44 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE UP | 104 | 67 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 589 | 595 | 1A | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU | ||||
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-1/206 | 3512 | 3518 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-124.1 | 1420 | 1426 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 1420 | 1426 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-125/351 | 1279 | 1285 | 1A | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-128 | 676 | 682 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU | ||||
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-130/301 | 2853 | 2860 | 1A,m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-138 | 3266 | 3272 | 1A | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-141/200a | 2689 | 2695 | m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG | ||||
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-142-3p | 1725 | 1732 | 1A,m8 | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-145 | 970 | 976 | 1A | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU | ||||
| miR-15/16/195/424/497 | 47 | 53 | 1A | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG | ||||
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG | ||||
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG | ||||
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-150 | 153 | 159 | 1A | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-153 | 1312 | 1318 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-155 | 559 | 565 | m8 | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-181 | 489 | 495 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-183 | 1170 | 1176 | m8 | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-185 | 265 | 271 | 1A | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-188 | 1281 | 1287 | 1A | hsa-miR-188 | CAUCCCUUGCAUGGUGGAGGGU |
| miR-192/215 | 1797 | 1804 | 1A,m8 | hsa-miR-192 | CUGACCUAUGAAUUGACAGCC |
| hsa-miR-215 | AUGACCUAUGAAUUGACAGAC | ||||
| miR-193 | 1591 | 1597 | 1A | hsa-miR-193a | AACUGGCCUACAAAGUCCCAG |
| hsa-miR-193b | AACUGGCCCUCAAAGUCCCGCUUU | ||||
| miR-196 | 2341 | 2347 | m8 | hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGG |
| hsa-miR-196b | UAGGUAGUUUCCUGUUGUUGG | ||||
| miR-199 | 968 | 975 | 1A,m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-200bc/429 | 1593 | 1600 | 1A,m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-203.1 | 398 | 404 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-21 | 52 | 58 | 1A | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA | ||||
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-217 | 3284 | 3290 | m8 | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-223 | 312 | 319 | 1A,m8 | hsa-miR-223 | UGUCAGUUUGUCAAAUACCCC |
| miR-27 | 1682 | 1689 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-29 | 2544 | 2550 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-30-5p | 2401 | 2407 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-362 | 275 | 281 | m8 | hsa-miR-362 | AAUCCUUGGAACCUAGGUGUGAGU |
| miR-369-3p | 1595 | 1601 | m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 612 | 618 | 1A | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-378 | 1095 | 1101 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| miR-380-5p | 3263 | 3269 | 1A | hsa-miR-380-5p | UGGUUGACCAUAGAACAUGCGC |
| hsa-miR-563 | AGGUUGACAUACGUUUCCC | ||||
| miR-381 | 2729 | 2735 | m8 | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-409-5p | 1330 | 1337 | 1A,m8 | hsa-miR-409-5p | AGGUUACCCGAGCAACUUUGCA |
| miR-448 | 1311 | 1318 | 1A,m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-493-5p | 3486 | 3492 | 1A | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-495 | 3307 | 3313 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-503 | 47 | 53 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-543 | 2328 | 2334 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| miR-96 | 715 | 721 | m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.