Gene Page: BCAR1
Summary ?
| GeneID | 9564 |
| Symbol | BCAR1 |
| Synonyms | CAS|CAS1|CASS1|CRKAS|P130Cas |
| Description | breast cancer anti-estrogen resistance 1 |
| Reference | MIM:602941|HGNC:HGNC:971|Ensembl:ENSG00000050820|HPRD:04248|Vega:OTTHUMG00000137604 |
| Gene type | protein-coding |
| Map location | 16q23.1 |
| Pascal p-value | 0.13 |
| Sherlock p-value | 0.531 |
| Fetal beta | -0.467 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| Expression | Meta-analysis of gene expression | P value: 2.094 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04914057 | 16 | 75283093 | BCAR1 | 1.729E-4 | -0.306 | 0.033 | DMG:Wockner_2014 |
| cg02438653 | 16 | 75285726 | BCAR1 | 2.66E-8 | -0.012 | 8.45E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9592471 | chr13 | 34989794 | BCAR1 | 9564 | 0.1 | trans |
Section II. Transcriptome annotation
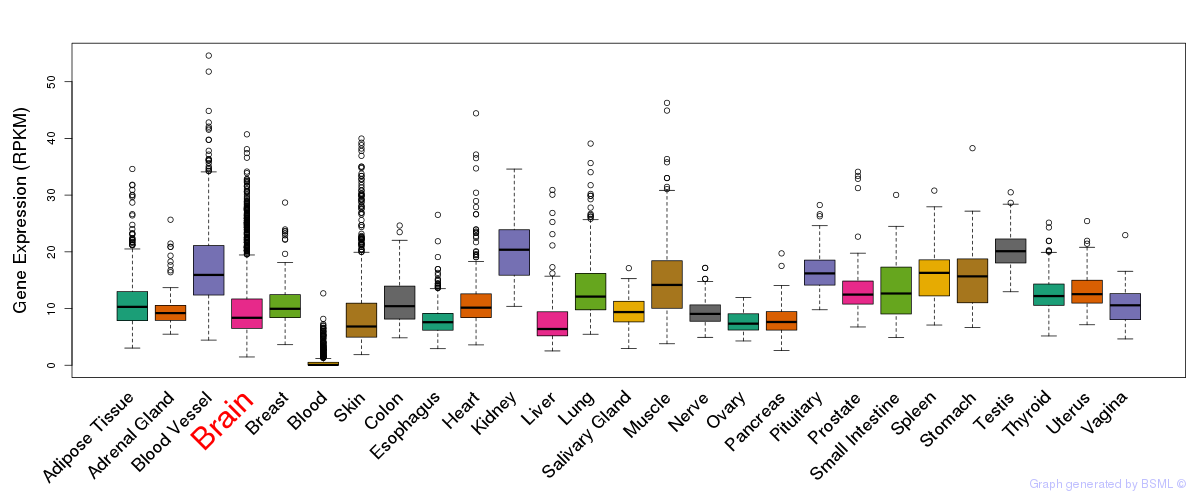
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | TAS | 8070403 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 9748234 |10339567 |10587647 |11782456 |14688263 |16105984 | |
| GO:0017124 | SH3 domain binding | IEA | - | |
| GO:0019903 | protein phosphatase binding | IPI | 9285683 | |
| GO:0019901 | protein kinase binding | IPI | 8070403 |8649368 |9020138 |9472046 |10026197 |16245368 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001558 | regulation of cell growth | TAS | 11607844 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | ISS | - | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | NAS | 10092204 | |
| GO:0007173 | epidermal growth factor receptor signaling pathway | ISS | - | |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0007229 | integrin-mediated signaling pathway | IDA | 8649368 | |
| GO:0007229 | integrin-mediated signaling pathway | ISS | - | |
| GO:0008283 | cell proliferation | TAS | 10639512 | |
| GO:0008286 | insulin receptor signaling pathway | ISS | - | |
| GO:0048011 | nerve growth factor receptor signaling pathway | ISS | - | |
| GO:0048008 | platelet-derived growth factor receptor signaling pathway | ISS | - | |
| GO:0007015 | actin filament organization | IDA | 16105984 | |
| GO:0007015 | actin filament organization | ISS | - | |
| GO:0042981 | regulation of apoptosis | TAS | 11607844 | |
| GO:0050852 | T cell receptor signaling pathway | NAS | 9295052 | |
| GO:0051301 | cell division | NAS | 10675905 | |
| GO:0030335 | positive regulation of cell migration | IDA | 9472046 | |
| GO:0030335 | positive regulation of cell migration | ISS | - | |
| GO:0050853 | B cell receptor signaling pathway | IDA | 9020138 | |
| GO:0050853 | B cell receptor signaling pathway | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001726 | ruffle | IDA | 9472046 | |
| GO:0001726 | ruffle | ISS | - | |
| GO:0005624 | membrane fraction | IDA | 8070403 | |
| GO:0005624 | membrane fraction | ISS | - | |
| GO:0005737 | cytoplasm | IDA | 8070403 | |
| GO:0005737 | cytoplasm | ISS | - | |
| GO:0030027 | lamellipodium | IEA | - | |
| GO:0005925 | focal adhesion | ISS | - | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD,BioGRID | 7780740 |
| BCAR3 | KIAA0554 | NSP2 | SH2D3B | breast cancer anti-estrogen resistance 3 | Affinity Capture-Western | BioGRID | 12517963 |
| CD2AP | CMS | DKFZp586H0519 | CD2-associated protein | - | HPRD,BioGRID | 10339567 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | Affinity Capture-Western Reconstituted Complex | BioGRID | 9581808 |9920935 |10085298 |10329689 |11369773 |12615911 |14657239 |15492270 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | - | HPRD,BioGRID | 10753828 |
| CSPG4 | HMW-MAA | MCSP | MCSPG | MEL-CSPG | MSK16 | NG2 | chondroitin sulfate proteoglycan 4 | - | HPRD | 10587647 |
| DOCK1 | DOCK180 | ced5 | dedicator of cytokinesis 1 | - | HPRD,BioGRID | 12615911 |
| E2F2 | E2F-2 | E2F transcription factor 2 | Two-hybrid | BioGRID | 16169070 |
| EFS | CAS3 | CASS3 | EFS1 | EFS2 | HEFS | SIN | embryonal Fyn-associated substrate | Two-hybrid | BioGRID | 16169070 |
| FES | FPS | feline sarcoma oncogene | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 8999909 |
| FXYD6 | - | FXYD domain containing ion transport regulator 6 | Two-hybrid | BioGRID | 16169070 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 9188452 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 8649368 |8700507 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10085298 |10822386 |
| GSN | DKFZp313L0718 | gelsolin (amyloidosis, Finnish type) | - | HPRD | 11577104 |
| HCK | JTK9 | hemopoietic cell kinase | - | HPRD,BioGRID | 9020138 |
| HSPA5 | BIP | FLJ26106 | GRP78 | MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | Two-hybrid | BioGRID | 16169070 |
| ID2 | GIG8 | ID2A | ID2H | MGC26389 | bHLHb26 | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | Affinity Capture-Western | BioGRID | 10502414 |
| INPPL1 | SHIP2 | inositol polyphosphate phosphatase-like 1 | - | HPRD,BioGRID | 11158326 |
| JUB | Ajuba | MGC15563 | jub, ajuba homolog (Xenopus laevis) | - | HPRD | 15728191 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD,BioGRID | 9020138 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | - | HPRD,BioGRID | 9581808 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | Affinity Capture-Western | BioGRID | 11432831 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | - | HPRD,BioGRID | 12135674 |
| NEDD9 | CAS-L | CAS2 | CASL | CASS2 | HEF1 | dJ49G10.2 | dJ761I2.1 | neural precursor cell expressed, developmentally down-regulated 9 | - | HPRD | 8668148 |9366405 |
| NEDD9 | CAS-L | CAS2 | CASL | CASS2 | HEF1 | dJ49G10.2 | dJ761I2.1 | neural precursor cell expressed, developmentally down-regulated 9 | Affinity Capture-Western | BioGRID | 10502414 |
| NPHP1 | FLJ97602 | JBTS4 | NPH1 | SLSN1 | nephronophthisis 1 (juvenile) | - | HPRD,BioGRID | 10739664 |11493697 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD,BioGRID | 10799562 |
| PKD1 | PBP | polycystic kidney disease 1 (autosomal dominant) | - | HPRD | 11113628 |
| PPP1R15A | GADD34 | protein phosphatase 1, regulatory (inhibitor) subunit 15A | Two-hybrid | BioGRID | 16169070 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD | 11514617 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 8649427 |8810278 |11577104 |12135674 |12615911 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD | 7479864 |11514617 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | CAS interacts with FAK. This interaction was modeled on a demonstrated interaction between human CAS and mouse FAK. | BIND | 9148935 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 8995252 |
| PTPN1 | PTP1B | protein tyrosine phosphatase, non-receptor type 1 | - | HPRD,BioGRID | 8940134 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | Affinity Capture-Western | BioGRID | 9188452 |
| PTPN12 | PTP-PEST | PTPG1 | protein tyrosine phosphatase, non-receptor type 12 | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 8887669 |9285683 |9748319 |12714323 |
| PTPN12 | PTP-PEST | PTPG1 | protein tyrosine phosphatase, non-receptor type 12 | - | HPRD | 8887669 |9285683 |9748319 |9920935 |12714323 |
| PTPRF | FLJ43335 | FLJ45062 | FLJ45567 | LAR | protein tyrosine phosphatase, receptor type, F | - | HPRD | 10822386 |
| PTPRH | FLJ39938 | MGC133058 | MGC133059 | SAP-1 | protein tyrosine phosphatase, receptor type, H | - | HPRD,BioGRID | 11278335 |
| PXN | FLJ16691 | paxillin | Affinity Capture-Western | BioGRID | 8810278 |11577104 |
| RAPGEF1 | C3G | DKFZp781P1719 | GRF2 | Rap guanine nucleotide exchange factor (GEF) 1 | - | HPRD,BioGRID | 9748234 |
| RICS | GC-GAP | GRIT | KIAA0712 | MGC1892 | p200RhoGAP | p250GAP | Rho GTPase-activating protein | - | HPRD,BioGRID | 12446789 |12819203 |
| SFN | YWHAS | stratifin | Affinity Capture-MS | BioGRID | 15778465 |
| SH2D3A | NSP1 | SH2 domain containing 3A | - | HPRD,BioGRID | 10187783 |
| SH2D3C | CHAT | FLJ39664 | NSP3 | PRO34088 | SH2 domain containing 3C | Affinity Capture-Western | BioGRID | 10692442 |
| SH3KBP1 | CIN85 | GIG10 | MIG18 | SH3-domain kinase binding protein 1 | Affinity Capture-Western | BioGRID | 11071869 |
| SNIP | KIAA1684 | SNAP25-interacting protein | - | HPRD,BioGRID | 14657239 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | c-Src interacts with CAS. | BIND | 9148935 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10085298 |10487518 |10739664 |11514617 |12397603 |12615911 |
| TNS1 | DKFZp586K0617 | MGC88584 | MST091 | MST122 | MST127 | MSTP122 | MSTP127 | MXRA6 | TNS | tensin 1 | Affinity Capture-Western | BioGRID | 8810278 |
| TRIP6 | MGC10556 | MGC10558 | MGC29959 | MGC3837 | MGC4423 | OIP1 | ZRP-1 | thyroid hormone receptor interactor 6 | - | HPRD,BioGRID | 11782456 |14688263 |
| VCL | CMD1W | MVCL | vinculin | Affinity Capture-Western | BioGRID | 11577104 |
| VPS11 | END1 | PEP5 | RNF108 | hVPS11 | vacuolar protein sorting 11 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD,BioGRID | 10026197 |
| ZYX | ESP-2 | HED-2 | zyxin | Affinity Capture-Western | BioGRID | 11782456 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CELL2CELL PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CXCR4 PATHWAY | 24 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTEGRIN PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PYK2 PATHWAY | 31 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PTEN PATHWAY | 18 | 14 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| PID ENDOTHELIN PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID PRL SIGNALING EVENTS PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| PID LYSOPHOSPHOLIPID PATHWAY | 66 | 53 | All SZGR 2.0 genes in this pathway |
| PID ER NONGENOMIC PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID AVB3 OPN PATHWAY | 31 | 29 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| PID RET PATHWAY | 39 | 29 | All SZGR 2.0 genes in this pathway |
| PID NETRIN PATHWAY | 32 | 27 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A9B1 PATHWAY | 25 | 18 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID IGF1 PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID UPA UPAR PATHWAY | 42 | 30 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID EPHA2 FWD PATHWAY | 19 | 16 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A4B1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| PID RAC1 PATHWAY | 54 | 37 | All SZGR 2.0 genes in this pathway |
| PID FAK PATHWAY | 59 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | 15 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET AGGREGATION PLUG FORMATION | 36 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA UP | 92 | 57 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 9 | 92 | 59 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D DN | 64 | 35 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SUBTIL PROGESTIN TARGETS | 36 | 25 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |