Gene Page: MDC1
Summary ?
| GeneID | 9656 |
| Symbol | MDC1 |
| Synonyms | NFBD1 |
| Description | mediator of DNA damage checkpoint 1 |
| Reference | MIM:607593|HGNC:HGNC:21163|Ensembl:ENSG00000137337|HPRD:16252|Vega:OTTHUMG00000031075 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 4.552E-14 |
| Sherlock p-value | 0.902 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24700494 | 6 | 30684284 | MDC1 | 1.396E-4 | 0.278 | 0.031 | DMG:Wockner_2014 |
| cg05546064 | 6 | 30685074 | MDC1 | 8.55E-9 | -0.009 | 3.98E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10035168 | chr5 | 5974295 | MDC1 | 9656 | 0.13 | trans |
Section II. Transcriptome annotation
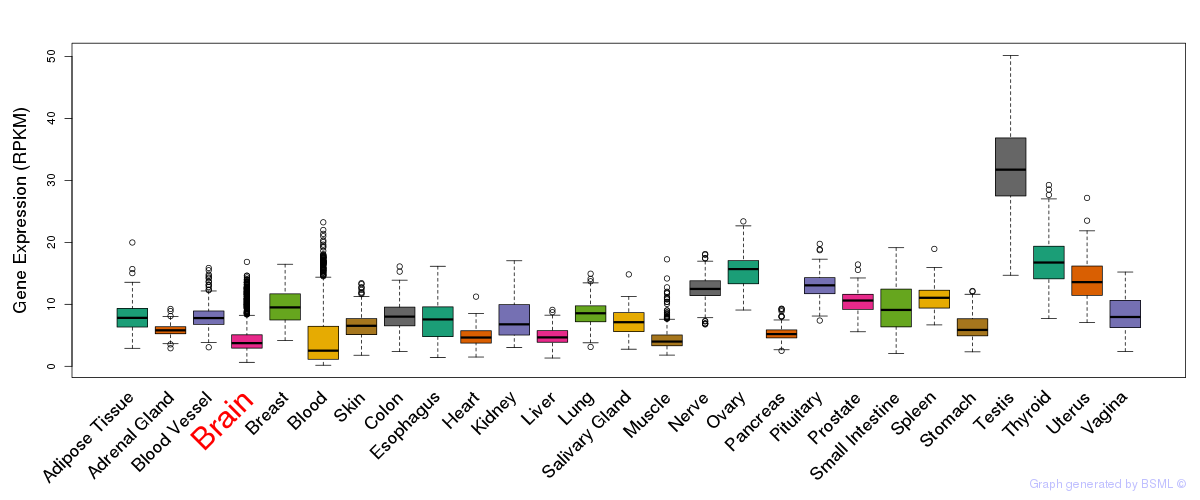
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 12607005 |17525332 |18001824 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006281 | DNA repair | IEA | - | |
| GO:0007049 | cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | IDA | 18029348 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IDA | 15604234 | |
| GO:0005654 | nucleoplasm | EXP | 12556884 |12607003 | |
| GO:0005925 | focal adhesion | IDA | 18029348 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | - | HPRD | 12607005 |
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | Affinity Capture-Western | BioGRID | 14519663 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD | 12607005 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Affinity Capture-Western | BioGRID | 17525332 |
| CENPC1 | CENP-C | CENPC | MIF2 | hcp-4 | centromere protein C 1 | Protein-peptide | BioGRID | 14578343 |
| CHEK2 | CDS1 | CHK2 | HuCds1 | LFS2 | PP1425 | RAD53 | CHK2 checkpoint homolog (S. pombe) | - | HPRD,BioGRID | 12551934 |12607004 |
| GATA4 | MGC126629 | GATA binding protein 4 | Protein-peptide | BioGRID | 14578343 |
| H2AFX | H2A.X | H2A/X | H2AX | H2A histone family, member X | - | HPRD,BioGRID | 12607005 |14519663 |
| HDAC10 | DKFZp761B039 | MGC149722 | histone deacetylase 10 | Protein-peptide | BioGRID | 14578343 |
| HDAC8 | HDACL1 | RPD3 | histone deacetylase 8 | Protein-peptide | BioGRID | 14578343 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Protein-peptide | BioGRID | 14578343 |
| MRE11A | ATLD | HNGS1 | MRE11 | MRE11B | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14519663 |
| NBN | AT-V1 | AT-V2 | ATV | FLJ10155 | MGC87362 | NBS | NBS1 | P95 | nibrin | - | HPRD,BioGRID | 12607003 |12607005 |14519663 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Protein-peptide | BioGRID | 14578343 |
| RAD50 | RAD50-2 | hRad50 | RAD50 homolog (S. cerevisiae) | Affinity Capture-Western | BioGRID | 14519663 |
| SMC1A | CDLS2 | DKFZp686L19178 | DXS423E | KIAA0178 | MGC138332 | SB1.8 | SMC1 | SMC1L1 | SMC1alpha | SMCB | structural maintenance of chromosomes 1A | - | HPRD | 12607005 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 14519663 |
| TP53BP1 | 53BP1 | FLJ41424 | MGC138366 | p202 | tumor protein p53 binding protein 1 | - | HPRD | 12607005 |
| WRN | DKFZp686C2056 | RECQ3 | RECQL2 | RECQL3 | Werner syndrome | Protein-peptide | BioGRID | 14578343 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ATM PATHWAY | 34 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | 17 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME DOUBLE STRAND BREAK REPAIR | 24 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION DN | 122 | 67 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC REGULATORS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| CROMER METASTASIS UP | 79 | 46 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN | 160 | 101 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 24HR | 128 | 73 | All SZGR 2.0 genes in this pathway |