Gene Page: REC8
Summary ?
| GeneID | 9985 |
| Symbol | REC8 |
| Synonyms | HR21spB|REC8L1|Rec8p |
| Description | REC8 meiotic recombination protein |
| Reference | MIM:608193|HGNC:HGNC:16879|Ensembl:ENSG00000100918|HPRD:09739|Vega:OTTHUMG00000171916 |
| Gene type | protein-coding |
| Map location | 14q12 |
| Pascal p-value | 0.154 |
| Sherlock p-value | 0.187 |
| Fetal beta | 2.191 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.047 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | REC8 | 9985 | 5.229E-8 | trans | ||
| rs3845734 | chr2 | 171125572 | REC8 | 9985 | 0 | trans | ||
| rs17762315 | chr5 | 76807576 | REC8 | 9985 | 0.07 | trans | ||
| rs16878317 | chr8 | 31657853 | REC8 | 9985 | 0.14 | trans | ||
| rs4075663 | chr11 | 18793398 | REC8 | 9985 | 0.15 | trans | ||
| rs16955618 | chr15 | 29937543 | REC8 | 9985 | 1.67E-11 | trans | ||
| rs1041786 | chr21 | 22617710 | REC8 | 9985 | 0.16 | trans |
Section II. Transcriptome annotation
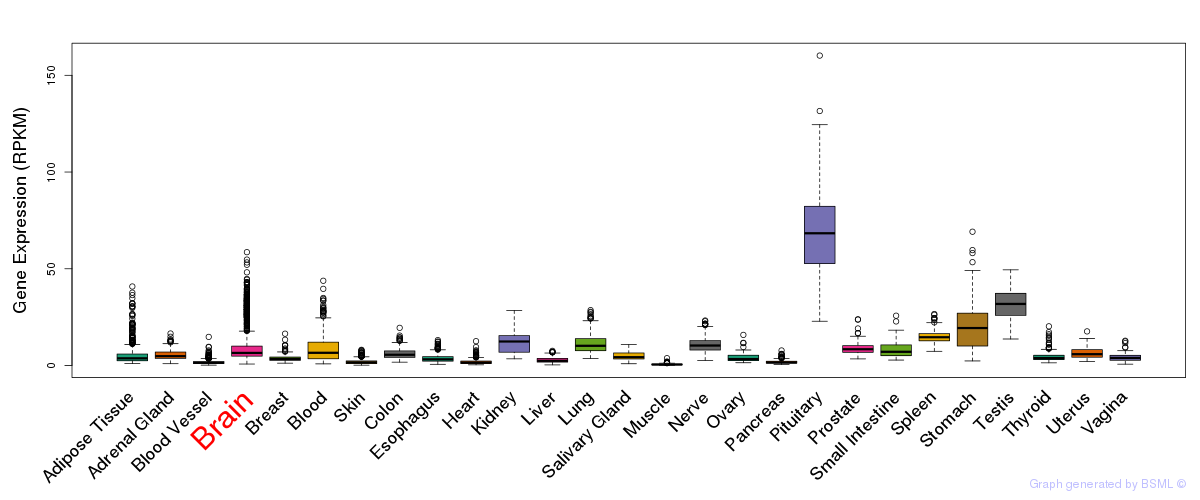
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| P4HB | 0.91 | 0.89 |
| COPB2 | 0.90 | 0.88 |
| PDIA4 | 0.89 | 0.90 |
| CALR | 0.89 | 0.87 |
| CSTF2T | 0.88 | 0.87 |
| SMU1 | 0.87 | 0.84 |
| G3BP1 | 0.87 | 0.86 |
| BZW1 | 0.87 | 0.87 |
| DDB1 | 0.87 | 0.85 |
| HSPD1 | 0.87 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.70 | -0.77 |
| AF347015.33 | -0.69 | -0.76 |
| AF347015.27 | -0.68 | -0.76 |
| FXYD1 | -0.68 | -0.75 |
| AF347015.31 | -0.68 | -0.76 |
| AF347015.8 | -0.68 | -0.76 |
| AF347015.21 | -0.67 | -0.72 |
| MT-CYB | -0.64 | -0.73 |
| AF347015.15 | -0.63 | -0.74 |
| AF347015.2 | -0.63 | -0.73 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007131 | reciprocal meiotic recombination | TAS | 10207075 | |
| GO:0007283 | spermatogenesis | TAS | 10207075 | |
| GO:0007062 | sister chromatid cohesion | TAS | 10207075 | |
| GO:0007126 | meiosis | TAS | 10207075 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000228 | nuclear chromosome | IEA | - | |
| GO:0000800 | lateral element | IEA | - | |
| GO:0005634 | nucleus | TAS | 10207075 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOSIS | 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CHROMOSOME MAINTENANCE | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC SYNAPSIS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| WANG HCP PROSTATE CANCER | 111 | 69 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP | 120 | 89 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR UP | 93 | 65 | All SZGR 2.0 genes in this pathway |
| ENGELMANN CANCER PROGENITORS DN | 70 | 44 | All SZGR 2.0 genes in this pathway |
| WEBER METHYLATED HCP IN FIBROBLAST DN | 42 | 22 | All SZGR 2.0 genes in this pathway |
| WEBER METHYLATED HCP IN SPERM UP | 20 | 13 | All SZGR 2.0 genes in this pathway |
| MATZUK MEIOTIC AND DNA REPAIR | 39 | 26 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOCYTE | 72 | 55 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3 UNMETHYLATED | 228 | 119 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN | 242 | 146 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS UP | 221 | 120 | All SZGR 2.0 genes in this pathway |