Application of DrVAEN in a breast cancer dataset (GEO ID: GSE20194)
1. Dataset summary
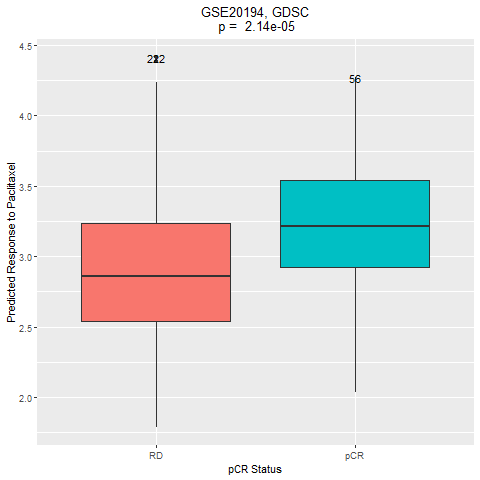
This dataset included 230 stage I-III breast cancers before any therapy. Gene expression data were generated from fine needle aspiration specimens using Affymetrix U133A microarrays. Patients received 6 months of chemotherapy including paclitaxel, 5-fluorouracil, cyclophosphamide and doxorubicin. The original study categorized patients as a pathological complete response (pCR = no residual invasive cancer in the breast or lymph nodes) or residual invasive cancer (RD). In our application, we compared the predicted response to paclitaxel (available in both the CCLE and GDSC panels) in the pCR group versus the RD group.
2. Preprocessing the dataset
The original gene expression data can be downloaded from GEO. Users may follow the code in GitHub to preprocess the data, including ID mapping and dealing with genes with multiple probe sets.
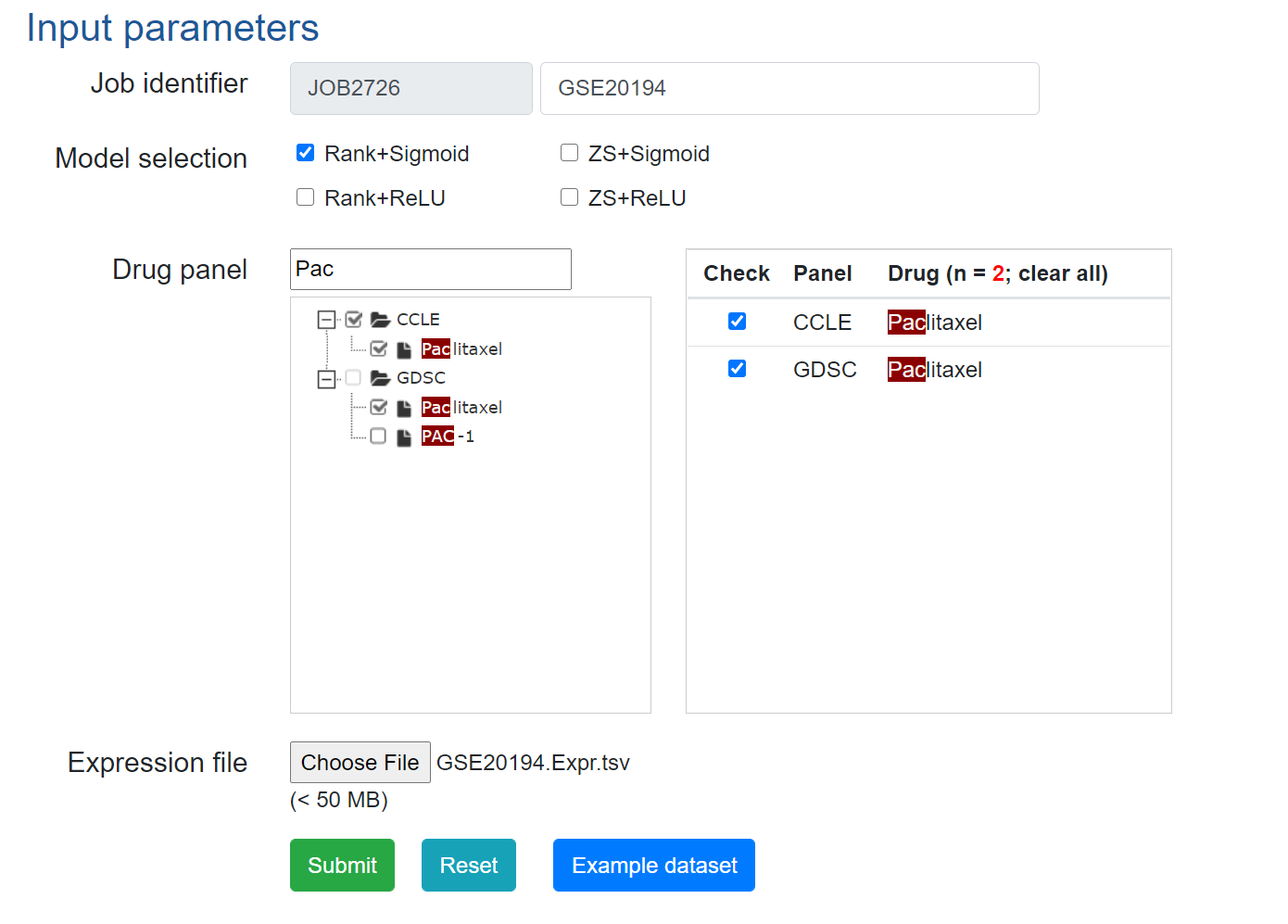
3. Predict drug responses using DrVAEN.
Example input parameters can be found below.
4. Comparation of the drug response between the two groups (pCR vs RD)