Application of DrVAEN in a breast cancer dataset (GEO ID: GSE25055)
1. Dataset summary
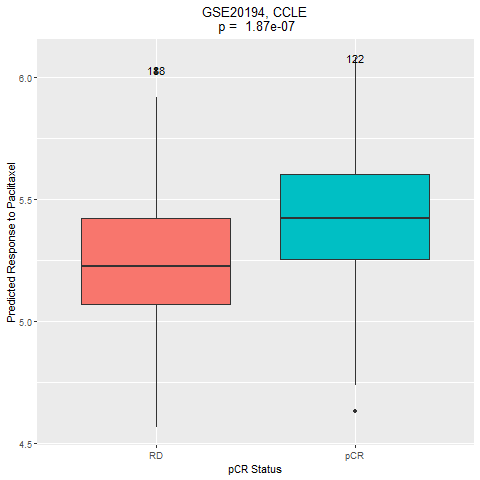
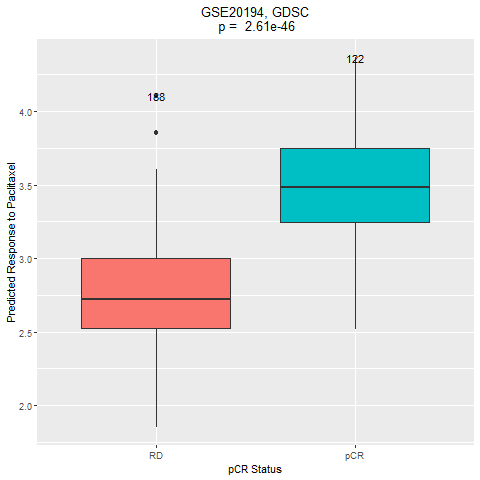
This dataset includes 310 HER2-negative breast cancer cases treated with taxane-anthracycline chemotherapy pre-operatively and endocrine therapy if ER-positive. Response was assessed at the end of neoadjuvant treatment. Samples were thus categorized into two groups: pCR and RD. The drug of interest is paclitaxel.
2. Get and prepare dataset
The original gene expression data can be downloaded from GEO. Users may follow the code in GitHub to preprocess the data, including ID mapping and dealing with genes with multiple probe sets.
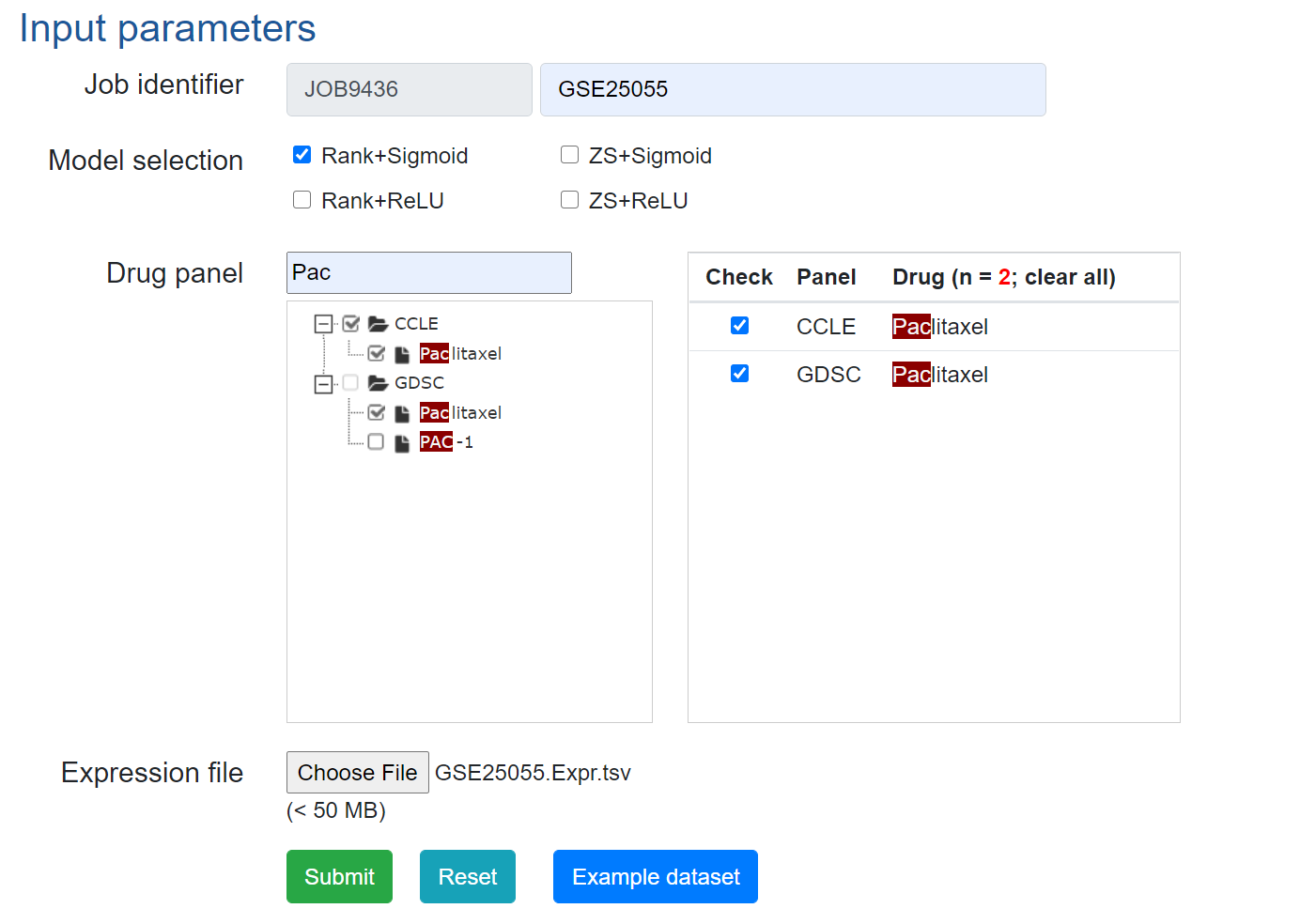
3. Predict drug responses using DrVAEN.
Example input parameters can be found below.
4. Comparation of the drug response between the two groups (pCR vs RD).