Application of DrVAEN in a non-small cell lung cancer dataset (GEO ID: GSE33072)
1. Dataset summary
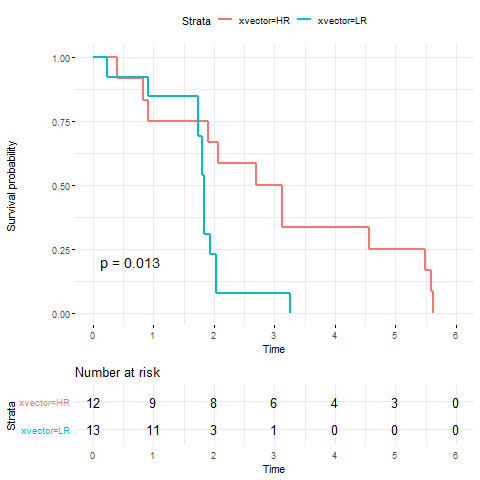
The original study aims at investigating the EMT gene signature in predicting resistance to EGFR inhibitors. Gene expression data for 131 core biopsies from patients with refractory non-small cell lung cancer (NSCLC) were generated using the Affymetrix Human Gene 1.0 ST Array. Among them, 25 patients also have their survival data available. We predicted the response to Erlotinib and stratified these samples based on the median of the predicted Erlotinib response. A survival analysis was conducted in the two groups.
2. Get and prepare dataset
The original gene expression data can be downloaded from GEO. Users may follow the code in GitHub to preprocess the data, including ID mapping and dealing with genes with multiple probe sets.
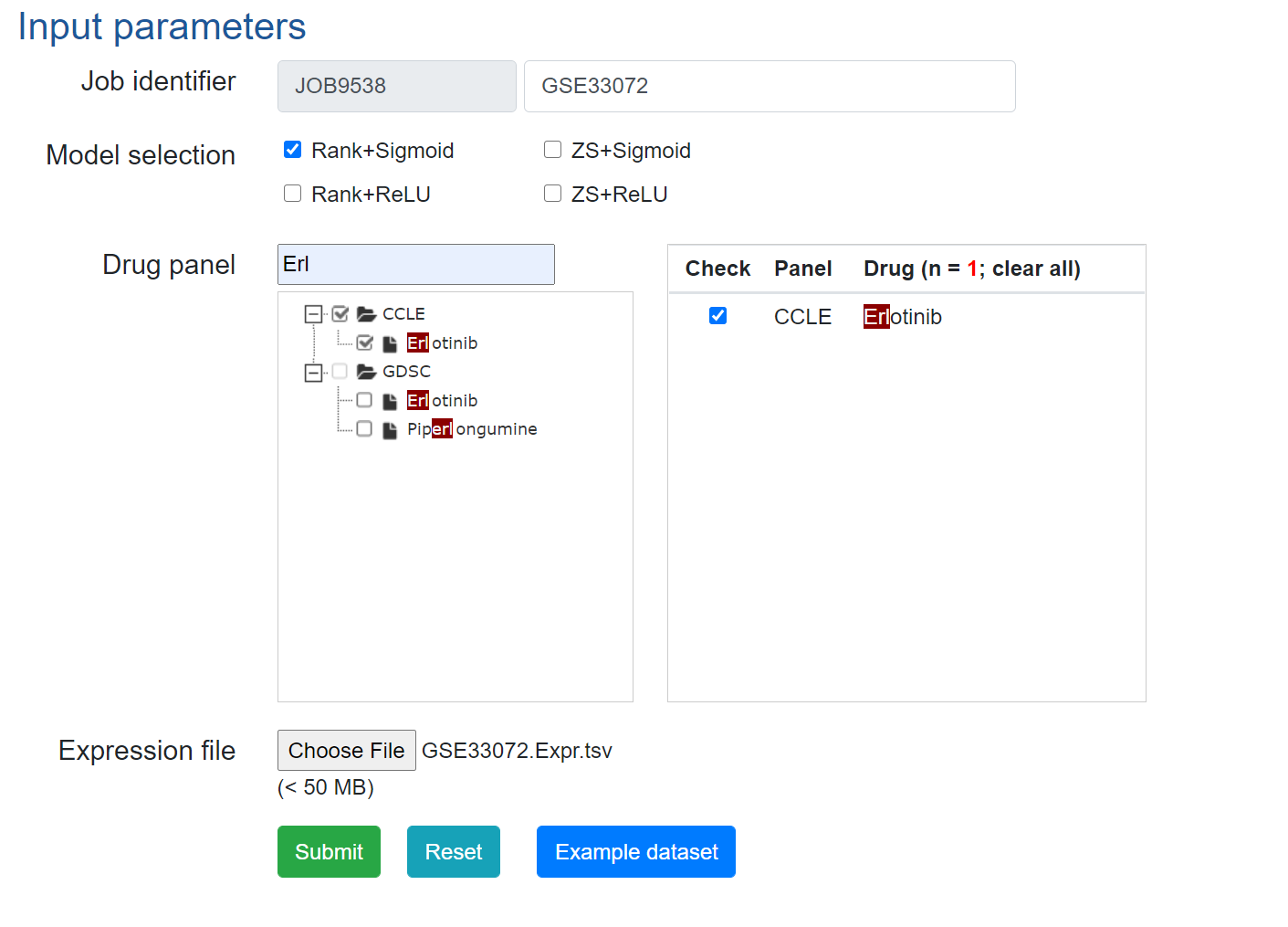
3. Predict drug responses using DrVAEN.
Example input parameters can be found below.
4. Survival analysis using the drug response data and survival data.