Genome-wide Association Studies
Genome-wide Association Studies
SNPs of Tier 1 set were collected from five independent resources: GWAS Catalog (CV:GWAScat), GWASdb, PheWAS, PGC2, and a multi-stage GWA study. These SNPs have strong evidence for their association with schizophrenia, reaching genome-wide significance in most cases. Raw records were remapped to the human reference genome hg19 if necessary. SNPs were then annotated systematically using the tool wANNOVAR. Raw records were displayed in the SNP page and the gene page in the same way as they were originally obtained.
SNPs of Tier 2 set were collected from the PGC2 summary results, with a nominal p-value <0.05. Tier 2 SNPs are searchable but not listed in the browse page because the number is too large.
| Study ID | # SNPs | # Genes | Tier | Reference | Inside | |
|---|---|---|---|---|---|---|
| CV:GWAScat | 560 | 486 | Tier 1 | NHGRI-EBI GWAS Catalog | SZGR 2.0 page for GWAScat | |
| CV:GWASdb | 560 | 2130 | Tier 1 | GWASdb | SZGR 2.0 page for GWASdb | |
| CV:PGC128 | 128 | 148 | Tier 1 | Psychiatric Genomics Consortium (PGC) | SZGR 2.0 page for PGC128 | |
| CV:PheWAS | 128 | 148 | Tier 1 | PheWAS | SZGR 2.0 page for PheWAS | |
| CV:Ripke_2013 | 128 | 27 | Tier 1 | PubMed | ||
| CV:PGCnp | 128 | 27 | Tier 2 | Psychiatric Genomics Consortium (PGC) | Link | |
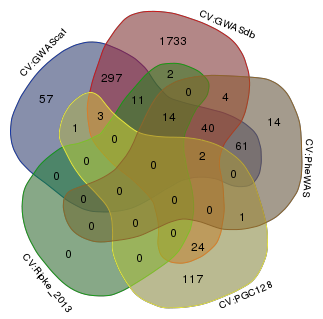
Venn diagram comparing the SNPs among the five Tier 1 SNPs.
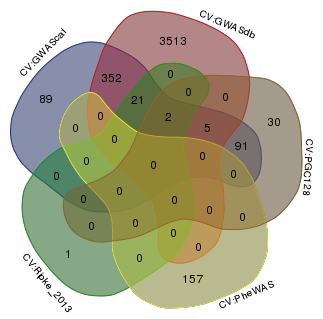
Venn diagram comparing the genes mapped to Tier 1 SNPs within 50kb.