Gene Page: ABI1
Summary ?
| GeneID | 10006 |
| Symbol | ABI1 |
| Synonyms | ABI-1|ABLBP4|E3B1|NAP1BP|SSH3BP|SSH3BP1 |
| Description | abl interactor 1 |
| Reference | MIM:603050|HGNC:HGNC:11320|Ensembl:ENSG00000136754|HPRD:04336|Vega:OTTHUMG00000017848 |
| Gene type | protein-coding |
| Map location | 10p11.2 |
| Pascal p-value | 0.004 |
| Fetal beta | -0.74 |
| eGene | Myers' cis & trans Meta |
| Support | STRUCTURAL PLASTICITY G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2716734 | chr2 | 39947720 | ABI1 | 10006 | 1.834E-6 | trans | ||
| rs2716736 | chr2 | 39947946 | ABI1 | 10006 | 1.834E-6 | trans | ||
| rs6446391 | chr4 | 5695478 | ABI1 | 10006 | 0.17 | trans | ||
| rs17023105 | chr4 | 96074164 | ABI1 | 10006 | 0.01 | trans | ||
| rs10516959 | chr4 | 96088848 | ABI1 | 10006 | 0.11 | trans | ||
| rs16869878 | chr5 | 2523760 | ABI1 | 10006 | 0.05 | trans | ||
| rs467856 | chr5 | 3128168 | ABI1 | 10006 | 0.11 | trans | ||
| snp_a-2169623 | 0 | ABI1 | 10006 | 1.694E-5 | trans | |||
| rs13175086 | chr5 | 174691428 | ABI1 | 10006 | 0.11 | trans | ||
| rs17165806 | chr7 | 93469599 | ABI1 | 10006 | 0.05 | trans | ||
| rs7777209 | chr7 | 93485280 | ABI1 | 10006 | 0.05 | trans | ||
| snp_a-2308001 | 0 | ABI1 | 10006 | 0.08 | trans | |||
| rs870686 | chr8 | 25253521 | ABI1 | 10006 | 0.02 | trans | ||
| rs10503771 | chr8 | 25304019 | ABI1 | 10006 | 0.02 | trans | ||
| rs12255258 | chr10 | 106306927 | ABI1 | 10006 | 0.06 | trans | ||
| rs11601862 | chr11 | 76333269 | ABI1 | 10006 | 0.2 | trans | ||
| rs7298298 | chr12 | 17810638 | ABI1 | 10006 | 0.13 | trans | ||
| rs2764585 | chr13 | 46880234 | ABI1 | 10006 | 0.19 | trans | ||
| rs9590075 | chr13 | 95414389 | ABI1 | 10006 | 0.02 | trans | ||
| rs1507047 | chr16 | 50932778 | ABI1 | 10006 | 0.18 | trans | ||
| rs349973 | chr17 | 50642288 | ABI1 | 10006 | 0.01 | trans | ||
| rs6128541 | chr20 | 57917688 | ABI1 | 10006 | 0.2 | trans |
Section II. Transcriptome annotation
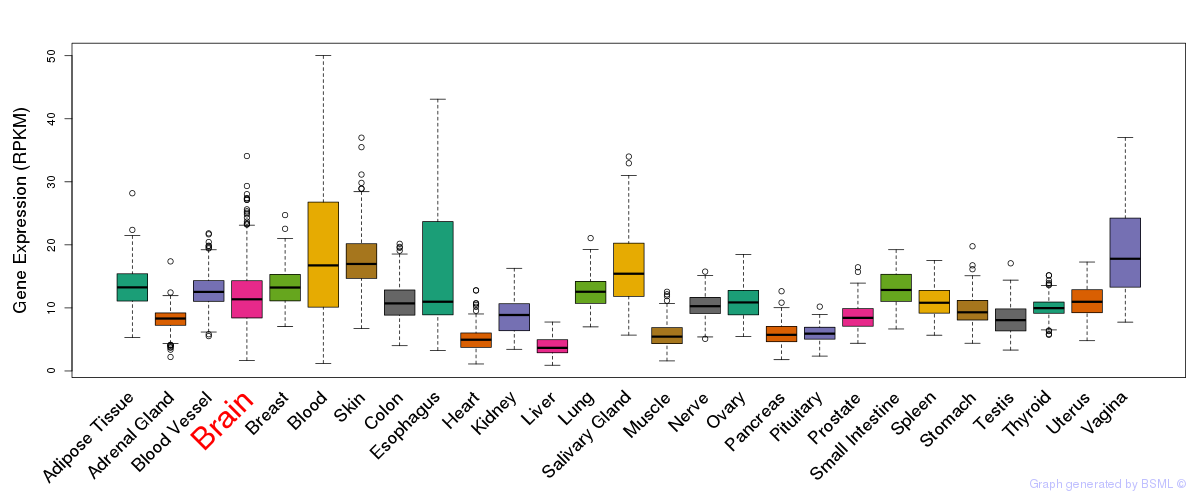
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008092 | cytoskeletal protein binding | TAS | 9593709 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | TAS | 9010225 | |
| GO:0008285 | negative regulation of cell proliferation | TAS | 9010225 | |
| GO:0018108 | peptidyl-tyrosine phosphorylation | IDA | 17101133 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005829 | cytosol | TAS | 9010225 | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005622 | intracellular | IDA | 17101133 | |
| GO:0005625 | soluble fraction | TAS | 9010225 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005783 | endoplasmic reticulum | TAS | 9593709 | |
| GO:0030027 | lamellipodium | IDA | 17101133 | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD | 9010225 |12672821 |
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | hNap1BP interacts with Abl. | BIND | 11418237 |
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | Affinity Capture-Western Reconstituted Complex | BioGRID | 9010225 |11418237 |12672821 |
| DNM2 | CMTDI1 | CMTDIB | DI-CMTB | DYN2 | DYNII | dynamin 2 | An unspecified isoform of Dyn2 interacts with Abi-1. This interaction was modeled on a demonstrated interaction between human Dyn2 and Abi-1 from an unspecified species. | BIND | 15696170 |
| ENAH | ENA | MENA | NDPP1 | enabled homolog (Drosophila) | - | HPRD,BioGRID | 12672821 |
| EPS8 | - | epidermal growth factor receptor pathway substrate 8 | - | HPRD | 1049958 |9010225 |11099046 |12127568 |
| EPS8 | - | epidermal growth factor receptor pathway substrate 8 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9010225 |11099046 |14565974 |
| EPS8L1 | DRC3 | EPS8R1 | FLJ20258 | MGC23164 | MGC4642 | PP10566 | EPS8-like 1 | - | HPRD,BioGRID | 14565974 |
| EPS8L2 | EPS8R2 | FLJ16738 | FLJ21935 | FLJ22171 | MGC126530 | MGC3088 | EPS8-like 2 | Affinity Capture-Western | BioGRID | 14565974 |
| EPS8L3 | EPS8R3 | FLJ21522 | MGC16817 | EPS8-like 3 | Affinity Capture-Western | BioGRID | 14565974 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD | 15048123 |
| NCF1C | SH3PXD1C | neutrophil cytosolic factor 1C pseudogene | ABI-1 interacts with p47phox. | BIND | 12681507 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | - | HPRD,BioGRID | 11418237 |
| NCK2 | GRB4 | NCKbeta | NCK adaptor protein 2 | - | HPRD | 11557983 |
| NCKAP1 | FLJ11291 | HEM2 | KIAA0587 | MGC8981 | NAP1 | NAP125 | NCK-associated protein 1 | Affinity Capture-Western Two-hybrid | BioGRID | 11418237 |
| NCKAP1 | FLJ11291 | HEM2 | KIAA0587 | MGC8981 | NAP1 | NAP125 | NCK-associated protein 1 | Nap1 interacts with hNap1BP. | BIND | 11418237 |
| NIN | KIAA1565 | ninein (GSK3B interacting protein) | - | HPRD | 15048123 |
| PAK2 | PAK65 | PAKgamma | p21 protein (Cdc42/Rac)-activated kinase 2 | - | HPRD | 11956071 |
| SOS1 | GF1 | GGF1 | GINGF | HGF | NS4 | son of sevenless homolog 1 (Drosophila) | - | HPRD,BioGRID | 10499589 |
| SOS2 | FLJ25596 | son of sevenless homolog 2 (Drosophila) | - | HPRD | 11003655 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ECADHERIN NASCENT AJ PATHWAY | 39 | 33 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| PID RAC1 REG PATHWAY | 38 | 25 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A4B1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| PID RAC1 PATHWAY | 54 | 37 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| XU AKT1 TARGETS 6HR | 27 | 18 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS UP | 50 | 30 | All SZGR 2.0 genes in this pathway |
| CHANG POU5F1 TARGETS UP | 15 | 10 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-181 | 131 | 137 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.