Gene Page: CWC27
Summary ?
| GeneID | 10283 |
| Symbol | CWC27 |
| Synonyms | NY-CO-10|SDCCAG-10|SDCCAG10 |
| Description | CWC27 spliceosome associated protein homolog |
| Reference | HGNC:HGNC:10664|Ensembl:ENSG00000153015|HPRD:11541|Vega:OTTHUMG00000074069 |
| Gene type | protein-coding |
| Map location | 5q12.3 |
| Pascal p-value | 0.062 |
| Sherlock p-value | 0.641 |
| Fetal beta | 0.067 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
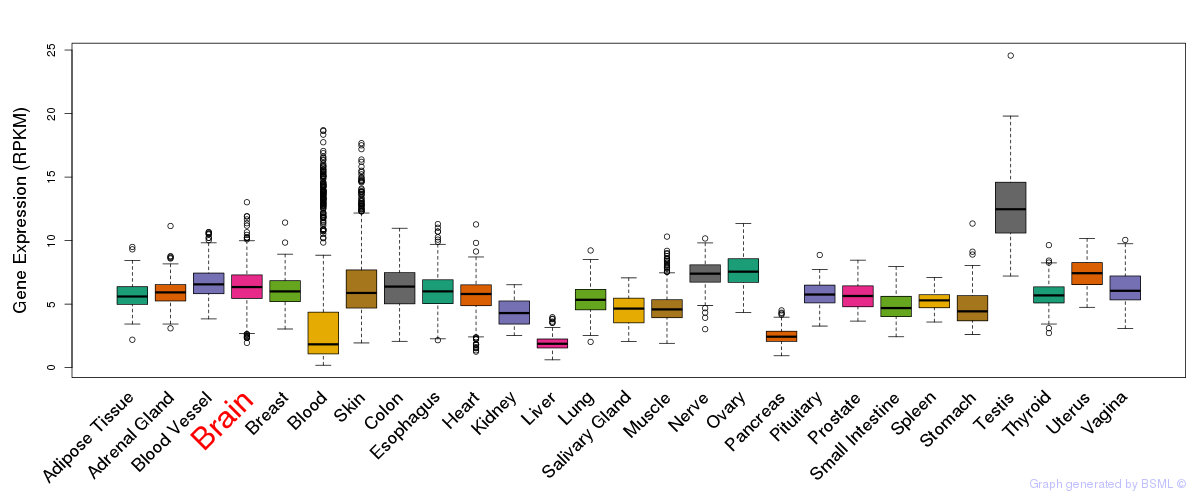
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KRTCAP2 | 0.86 | 0.85 |
| PSMB6 | 0.83 | 0.76 |
| MRPL36 | 0.82 | 0.78 |
| C11orf59 | 0.81 | 0.82 |
| NDUFS8 | 0.81 | 0.81 |
| CUTA | 0.81 | 0.77 |
| HDHD3 | 0.81 | 0.79 |
| RABAC1 | 0.80 | 0.78 |
| ANAPC11 | 0.80 | 0.83 |
| NDUFA11 | 0.80 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| Z83840.4 | -0.42 | -0.45 |
| AC010300.1 | -0.41 | -0.50 |
| MT-ATP8 | -0.41 | -0.37 |
| AF347015.18 | -0.41 | -0.37 |
| AP000769.2 | -0.39 | -0.47 |
| EIF5B | -0.38 | -0.46 |
| SEC62 | -0.37 | -0.45 |
| AC016705.1 | -0.37 | -0.37 |
| AF347015.26 | -0.35 | -0.32 |
| TRIM44 | -0.35 | -0.33 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING DN | 35 | 24 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| EHLERS ANEUPLOIDY UP | 41 | 29 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |