Gene Page: SF3A1
Summary ?
| GeneID | 10291 |
| Symbol | SF3A1 |
| Synonyms | PRP21|PRPF21|SAP114|SF3A120 |
| Description | splicing factor 3a subunit 1 |
| Reference | MIM:605595|HGNC:HGNC:10765|Ensembl:ENSG00000099995|HPRD:06903|Vega:OTTHUMG00000151005 |
| Gene type | protein-coding |
| Map location | 22q12.2 |
| Pascal p-value | 0.002 |
| Sherlock p-value | 0.032 |
| Fetal beta | -0.096 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11475510 | 22 | 30752584 | SF3A1 | 2.14E-8 | -0.008 | 7.26E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12408656 | chr1 | 162946658 | SF3A1 | 10291 | 0.18 | trans |
Section II. Transcriptome annotation
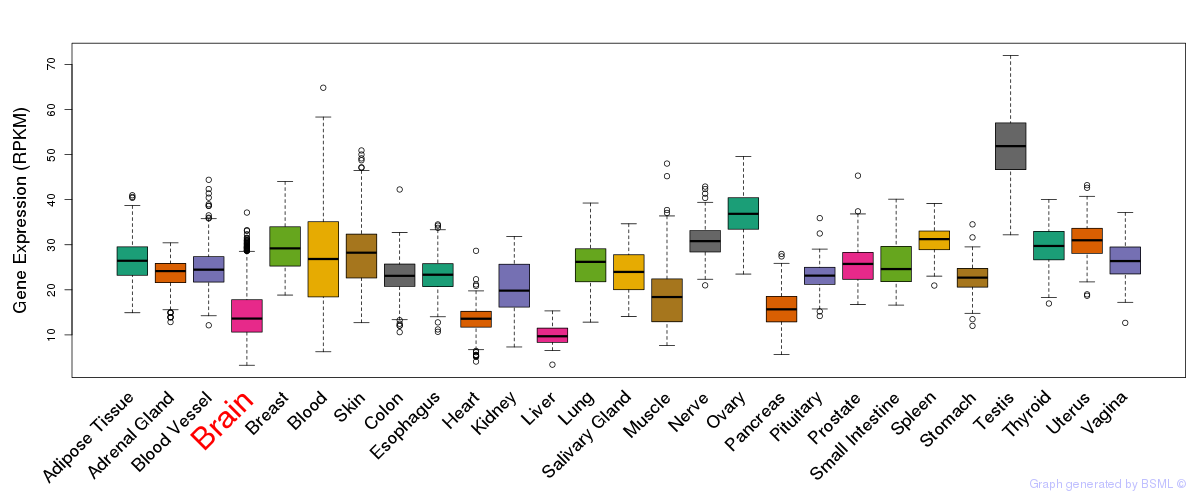
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TCEAL7 | 0.78 | 0.85 |
| CALM2 | 0.76 | 0.84 |
| UBE2B | 0.74 | 0.75 |
| GLRX2 | 0.74 | 0.77 |
| NKIRAS1 | 0.73 | 0.81 |
| MOCS2 | 0.72 | 0.76 |
| COMMD8 | 0.72 | 0.79 |
| KRT222P | 0.72 | 0.73 |
| NDUFAF4 | 0.71 | 0.80 |
| FBXO9 | 0.71 | 0.76 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HIF3A | -0.47 | -0.54 |
| AC020763.2 | -0.45 | -0.49 |
| CDC42EP4 | -0.44 | -0.50 |
| SEMA4B | -0.43 | -0.47 |
| FAM59B | -0.43 | -0.48 |
| RP4-697K14.1 | -0.43 | -0.53 |
| SMTN | -0.43 | -0.51 |
| EPHA2 | -0.42 | -0.50 |
| MTSS1L | -0.39 | -0.44 |
| NFKB2 | -0.39 | -0.45 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003723 | RNA binding | IDA | 10882114 | |
| GO:0003723 | RNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 10976766 |17353931 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000389 | nuclear mRNA 3'-splice site recognition | TAS | 15647371 | |
| GO:0000398 | nuclear mRNA splicing, via spliceosome | EXP | 12226669 | |
| GO:0000398 | nuclear mRNA splicing, via spliceosome | IDA | 10882114 | |
| GO:0006396 | RNA processing | IEA | - | |
| GO:0008380 | RNA splicing | IEA | - | |
| GO:0006464 | protein modification process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005684 | U2-dependent spliceosome | IDA | 10882114 | |
| GO:0005730 | nucleolus | IDA | 18029348 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING | 111 | 58 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN | 186 | 114 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS | 77 | 55 | All SZGR 2.0 genes in this pathway |
| DER IFN GAMMA RESPONSE UP | 71 | 45 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP | 113 | 71 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| THILLAINADESAN ZNF217 TARGETS UP | 44 | 22 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 DN | 180 | 116 | All SZGR 2.0 genes in this pathway |