Gene Page: BCKDK
Summary ?
| GeneID | 10295 |
| Symbol | BCKDK |
| Synonyms | BCKDKD|BDK |
| Description | branched chain ketoacid dehydrogenase kinase |
| Reference | MIM:614901|HGNC:HGNC:16902|Ensembl:ENSG00000103507|HPRD:12523|Vega:OTTHUMG00000047356 |
| Gene type | protein-coding |
| Map location | 16p11.2 |
| Pascal p-value | 0.174 |
| Sherlock p-value | 0.872 |
| Fetal beta | -0.643 |
| DMG | 1 (# studies) |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_mitochondria G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15785580 | 16 | 31119650 | BCKDK | -0.02 | 0.51 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
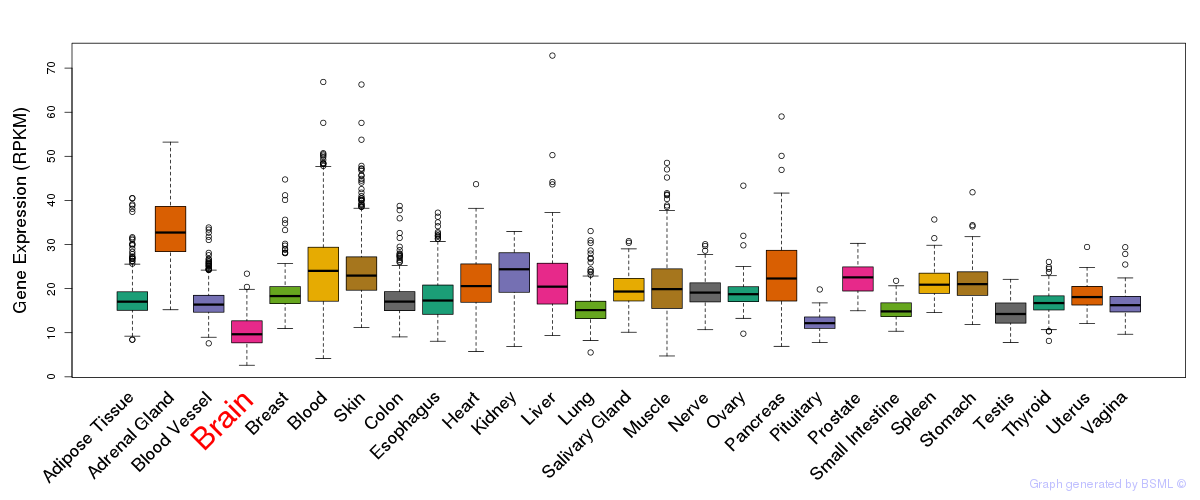
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000155 | two-component sensor activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005524 | ATP binding | ISS | - | |
| GO:0004674 | protein serine/threonine kinase activity | ISS | - | |
| GO:0016301 | kinase activity | IEA | - | |
| GO:0047323 | [3-methyl-2-oxobutanoate dehydrogenase (acetyl-transferring)] kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0009083 | branched chain family amino acid catabolic process | ISS | - | |
| GO:0009083 | branched chain family amino acid catabolic process | TAS | 11839747 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0016310 | phosphorylation | ISS | - | |
| GO:0016310 | phosphorylation | TAS | 11839747 | |
| GO:0018106 | peptidyl-histidine phosphorylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005739 | mitochondrion | TAS | 11839747 | |
| GO:0005759 | mitochondrial matrix | IEA | - | |
| GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex | ISS | - | |
| GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex | TAS | 11839747 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCKDHA | BCKDE1A | FLJ45695 | MSU | MSUD1 | OVD1A | branched chain keto acid dehydrogenase E1, alpha polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| BCKDHB | E1B | FLJ17880 | dJ279A18.1 | branched chain keto acid dehydrogenase E1, beta polypeptide | - | HPRD | 1377677 |
| CAB39 | CGI-66 | FLJ22682 | MO25 | calcium binding protein 39 | Affinity Capture-MS | BioGRID | 17353931 |
| CAB39L | FLJ12577 | MO25-BETA | MO2L | RP11-103J18.3 | bA103J18.3 | calcium binding protein 39-like | Affinity Capture-MS | BioGRID | 17353931 |
| DBT | BCATE2 | E2 | E2B | MGC9061 | dihydrolipoamide branched chain transacylase E2 | - | HPRD | 1377677 |
| DLD | DLDH | E3 | GCSL | LAD | PHE3 | dihydrolipoamide dehydrogenase | - | HPRD | 15576032 |
| FAM98B | FLJ38426 | family with sequence similarity 98, member B | Affinity Capture-MS | BioGRID | 17353931 |
| MUTED | DKFZp686E2287 | MU | muted homolog (mouse) | - | HPRD | 15102850 |
| RNF219 | C13orf7 | DKFZp686A01276 | DKFZp686N15250 | DKFZp686O03173 | FLJ13449 | FLJ25774 | ring finger protein 219 | Affinity Capture-MS | BioGRID | 17353931 |
| RTCD1 | RPC | RNA terminal phosphate cyclase domain 1 | Affinity Capture-MS | BioGRID | 17353931 |
| TRAF4 | CART1 | MLN62 | RNF83 | TNF receptor-associated factor 4 | TRAF4 interacts with BCKDK. | BIND | 12023963 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| LU IL4 SIGNALING | 94 | 56 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |