Gene Page: WARS2
Summary ?
| GeneID | 10352 |
| Symbol | WARS2 |
| Synonyms | TrpRS |
| Description | tryptophanyl tRNA synthetase 2, mitochondrial |
| Reference | MIM:604733|HGNC:HGNC:12730|Ensembl:ENSG00000116874|HPRD:05295|Vega:OTTHUMG00000012335 |
| Gene type | protein-coding |
| Map location | 1p12 |
| Pascal p-value | 0.867 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23043968 | 1 | 119682730 | WARS2 | 7.02E-8 | -0.017 | 1.7E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10923714 | chr1 | 119512912 | WARS2 | 10352 | 0.02 | cis | ||
| rs36781 | chr5 | 109289728 | WARS2 | 10352 | 0.09 | trans | ||
| rs7553422 | 1 | 119540719 | WARS2 | ENSG00000116874.7 | 2.395E-6 | 0.01 | 142575 | gtex_brain_ba24 |
| rs1886914 | 1 | 119541452 | WARS2 | ENSG00000116874.7 | 2.395E-6 | 0.01 | 141842 | gtex_brain_ba24 |
| rs10923724 | 1 | 119546842 | WARS2 | ENSG00000116874.7 | 4.36E-7 | 0.01 | 136452 | gtex_brain_ba24 |
| rs6428789 | 1 | 119550480 | WARS2 | ENSG00000116874.7 | 1.311E-6 | 0.01 | 132814 | gtex_brain_ba24 |
| rs2645303 | 1 | 119555146 | WARS2 | ENSG00000116874.7 | 6.319E-7 | 0.01 | 128148 | gtex_brain_ba24 |
| rs2765542 | 1 | 119560326 | WARS2 | ENSG00000116874.7 | 2.539E-6 | 0.01 | 122968 | gtex_brain_ba24 |
| rs2645305 | 1 | 119561365 | WARS2 | ENSG00000116874.7 | 2.539E-6 | 0.01 | 121929 | gtex_brain_ba24 |
| rs2645291 | 1 | 119572541 | WARS2 | ENSG00000116874.7 | 1.204E-6 | 0.01 | 110753 | gtex_brain_ba24 |
| rs12086 | 1 | 119573860 | WARS2 | ENSG00000116874.7 | 3.753E-6 | 0.01 | 109434 | gtex_brain_ba24 |
| rs2645294 | 1 | 119574587 | WARS2 | ENSG00000116874.7 | 3.435E-6 | 0.01 | 108707 | gtex_brain_ba24 |
| rs2794315 | 1 | 119577666 | WARS2 | ENSG00000116874.7 | 5.608E-7 | 0.01 | 105628 | gtex_brain_ba24 |
| rs2247883 | 1 | 119577837 | WARS2 | ENSG00000116874.7 | 4.558E-7 | 0.01 | 105457 | gtex_brain_ba24 |
| rs61808932 | 1 | 119643820 | WARS2 | ENSG00000116874.7 | 2.009E-6 | 0.01 | 39474 | gtex_brain_ba24 |
| rs3936018 | 1 | 119682370 | WARS2 | ENSG00000116874.7 | 1.196E-7 | 0.01 | 924 | gtex_brain_ba24 |
| rs55696509 | 1 | 119686656 | WARS2 | ENSG00000116874.7 | 1.196E-7 | 0.01 | -3362 | gtex_brain_ba24 |
| rs61590995 | 1 | 119699600 | WARS2 | ENSG00000116874.7 | 3.77E-6 | 0.01 | -16306 | gtex_brain_ba24 |
| rs72691108 | 1 | 119762175 | WARS2 | ENSG00000116874.7 | 1.615E-7 | 0.01 | -78881 | gtex_brain_ba24 |
| rs142454091 | 1 | 119476119 | WARS2 | ENSG00000116874.7 | 2.058E-6 | 0 | 207175 | gtex_brain_putamen_basal |
| rs7553422 | 1 | 119540719 | WARS2 | ENSG00000116874.7 | 2.148E-6 | 0 | 142575 | gtex_brain_putamen_basal |
| rs1886914 | 1 | 119541452 | WARS2 | ENSG00000116874.7 | 2.148E-6 | 0 | 141842 | gtex_brain_putamen_basal |
| rs6428789 | 1 | 119550480 | WARS2 | ENSG00000116874.7 | 2.248E-6 | 0 | 132814 | gtex_brain_putamen_basal |
| rs77152130 | 1 | 119551277 | WARS2 | ENSG00000116874.7 | 2.401E-8 | 0 | 132017 | gtex_brain_putamen_basal |
| rs2645303 | 1 | 119555146 | WARS2 | ENSG00000116874.7 | 5.303E-9 | 0 | 128148 | gtex_brain_putamen_basal |
| rs2765542 | 1 | 119560326 | WARS2 | ENSG00000116874.7 | 2.521E-7 | 0 | 122968 | gtex_brain_putamen_basal |
| rs2645305 | 1 | 119561365 | WARS2 | ENSG00000116874.7 | 2.544E-7 | 0 | 121929 | gtex_brain_putamen_basal |
| rs2794310 | 1 | 119561930 | WARS2 | ENSG00000116874.7 | 2.56E-7 | 0 | 121364 | gtex_brain_putamen_basal |
| rs2765543 | 1 | 119563411 | WARS2 | ENSG00000116874.7 | 9.097E-8 | 0 | 119883 | gtex_brain_putamen_basal |
| rs2645291 | 1 | 119572541 | WARS2 | ENSG00000116874.7 | 1.82E-8 | 0 | 110753 | gtex_brain_putamen_basal |
| rs12086 | 1 | 119573860 | WARS2 | ENSG00000116874.7 | 5.826E-9 | 0 | 109434 | gtex_brain_putamen_basal |
| rs1057991 | 1 | 119575170 | WARS2 | ENSG00000116874.7 | 1.482E-6 | 0 | 108124 | gtex_brain_putamen_basal |
| rs1057990 | 1 | 119575232 | WARS2 | ENSG00000116874.7 | 1.482E-6 | 0 | 108062 | gtex_brain_putamen_basal |
| rs2794315 | 1 | 119577666 | WARS2 | ENSG00000116874.7 | 1.194E-8 | 0 | 105628 | gtex_brain_putamen_basal |
| rs2247883 | 1 | 119577837 | WARS2 | ENSG00000116874.7 | 1.181E-8 | 0 | 105457 | gtex_brain_putamen_basal |
| rs11285792 | 1 | 119579945 | WARS2 | ENSG00000116874.7 | 1.193E-6 | 0 | 103349 | gtex_brain_putamen_basal |
| rs2794314 | 1 | 119581404 | WARS2 | ENSG00000116874.7 | 1.482E-6 | 0 | 101890 | gtex_brain_putamen_basal |
| rs2765529 | 1 | 119581900 | WARS2 | ENSG00000116874.7 | 1.482E-6 | 0 | 101394 | gtex_brain_putamen_basal |
| rs2645295 | 1 | 119582448 | WARS2 | ENSG00000116874.7 | 1.482E-6 | 0 | 100846 | gtex_brain_putamen_basal |
| rs1325938 | 1 | 119582642 | WARS2 | ENSG00000116874.7 | 1.631E-6 | 0 | 100652 | gtex_brain_putamen_basal |
| rs2765530 | 1 | 119584396 | WARS2 | ENSG00000116874.7 | 1.481E-6 | 0 | 98898 | gtex_brain_putamen_basal |
| rs2765531 | 1 | 119585497 | WARS2 | ENSG00000116874.7 | 1.479E-6 | 0 | 97797 | gtex_brain_putamen_basal |
| rs1325940 | 1 | 119586687 | WARS2 | ENSG00000116874.7 | 1.479E-6 | 0 | 96607 | gtex_brain_putamen_basal |
| rs2794313 | 1 | 119587440 | WARS2 | ENSG00000116874.7 | 1.479E-6 | 0 | 95854 | gtex_brain_putamen_basal |
| rs2645302 | 1 | 119590398 | WARS2 | ENSG00000116874.7 | 1.477E-6 | 0 | 92896 | gtex_brain_putamen_basal |
| rs2645301 | 1 | 119590420 | WARS2 | ENSG00000116874.7 | 1.477E-6 | 0 | 92874 | gtex_brain_putamen_basal |
| rs2794312 | 1 | 119591046 | WARS2 | ENSG00000116874.7 | 1.477E-6 | 0 | 92248 | gtex_brain_putamen_basal |
| rs2765532 | 1 | 119591546 | WARS2 | ENSG00000116874.7 | 1.477E-6 | 0 | 91748 | gtex_brain_putamen_basal |
| rs56708249 | 1 | 119593205 | WARS2 | ENSG00000116874.7 | 1.476E-6 | 0 | 90089 | gtex_brain_putamen_basal |
| rs17023171 | 1 | 119593268 | WARS2 | ENSG00000116874.7 | 1.476E-6 | 0 | 90026 | gtex_brain_putamen_basal |
| rs17023174 | 1 | 119593301 | WARS2 | ENSG00000116874.7 | 1.476E-6 | 0 | 89993 | gtex_brain_putamen_basal |
| rs12072640 | 1 | 119593644 | WARS2 | ENSG00000116874.7 | 3.159E-7 | 0 | 89650 | gtex_brain_putamen_basal |
| rs12073744 | 1 | 119593659 | WARS2 | ENSG00000116874.7 | 3.049E-8 | 0 | 89635 | gtex_brain_putamen_basal |
| rs12072768 | 1 | 119593918 | WARS2 | ENSG00000116874.7 | 3.159E-7 | 0 | 89376 | gtex_brain_putamen_basal |
| rs12077366 | 1 | 119593959 | WARS2 | ENSG00000116874.7 | 3.156E-7 | 0 | 89335 | gtex_brain_putamen_basal |
| rs7532510 | 1 | 119594236 | WARS2 | ENSG00000116874.7 | 2.772E-7 | 0 | 89058 | gtex_brain_putamen_basal |
| rs2794311 | 1 | 119595949 | WARS2 | ENSG00000116874.7 | 3.144E-7 | 0 | 87345 | gtex_brain_putamen_basal |
| rs2765533 | 1 | 119597395 | WARS2 | ENSG00000116874.7 | 2.627E-7 | 0 | 85899 | gtex_brain_putamen_basal |
| rs2765534 | 1 | 119597792 | WARS2 | ENSG00000116874.7 | 2.519E-7 | 0 | 85502 | gtex_brain_putamen_basal |
| rs144102861 | 1 | 119599159 | WARS2 | ENSG00000116874.7 | 1.776E-6 | 0 | 84135 | gtex_brain_putamen_basal |
| rs61808892 | 1 | 119599207 | WARS2 | ENSG00000116874.7 | 2.61E-7 | 0 | 84087 | gtex_brain_putamen_basal |
| rs12088921 | 1 | 119599970 | WARS2 | ENSG00000116874.7 | 2.104E-7 | 0 | 83324 | gtex_brain_putamen_basal |
| rs146862216 | 1 | 119600175 | WARS2 | ENSG00000116874.7 | 3.385E-8 | 0 | 83119 | gtex_brain_putamen_basal |
| rs10923740 | 1 | 119600287 | WARS2 | ENSG00000116874.7 | 3.433E-6 | 0 | 83007 | gtex_brain_putamen_basal |
| rs6668960 | 1 | 119600812 | WARS2 | ENSG00000116874.7 | 2.013E-7 | 0 | 82482 | gtex_brain_putamen_basal |
| rs6669254 | 1 | 119601091 | WARS2 | ENSG00000116874.7 | 8.48E-9 | 0 | 82203 | gtex_brain_putamen_basal |
| rs2361269 | 1 | 119602215 | WARS2 | ENSG00000116874.7 | 1.969E-7 | 0 | 81079 | gtex_brain_putamen_basal |
| rs2645300 | 1 | 119603067 | WARS2 | ENSG00000116874.7 | 1.952E-7 | 0 | 80227 | gtex_brain_putamen_basal |
| rs2361270 | 1 | 119603322 | WARS2 | ENSG00000116874.7 | 1.947E-7 | 0 | 79972 | gtex_brain_putamen_basal |
| rs2361271 | 1 | 119603405 | WARS2 | ENSG00000116874.7 | 1.947E-7 | 0 | 79889 | gtex_brain_putamen_basal |
| rs2361272 | 1 | 119603417 | WARS2 | ENSG00000116874.7 | 1.947E-7 | 0 | 79877 | gtex_brain_putamen_basal |
| rs2361273 | 1 | 119603700 | WARS2 | ENSG00000116874.7 | 1.959E-7 | 0 | 79594 | gtex_brain_putamen_basal |
| rs10923741 | 1 | 119604473 | WARS2 | ENSG00000116874.7 | 1.93E-7 | 0 | 78821 | gtex_brain_putamen_basal |
| rs10802073 | 1 | 119604655 | WARS2 | ENSG00000116874.7 | 1.93E-7 | 0 | 78639 | gtex_brain_putamen_basal |
| rs1343723 | 1 | 119605054 | WARS2 | ENSG00000116874.7 | 1.928E-7 | 0 | 78240 | gtex_brain_putamen_basal |
| rs138055448 | 1 | 119606819 | WARS2 | ENSG00000116874.7 | 1.917E-7 | 0 | 76475 | gtex_brain_putamen_basal |
| rs6695059 | 1 | 119606960 | WARS2 | ENSG00000116874.7 | 1.914E-7 | 0 | 76334 | gtex_brain_putamen_basal |
| rs17023185 | 1 | 119610787 | WARS2 | ENSG00000116874.7 | 2.183E-7 | 0 | 72507 | gtex_brain_putamen_basal |
| rs6662165 | 1 | 119614071 | WARS2 | ENSG00000116874.7 | 4.502E-9 | 0 | 69223 | gtex_brain_putamen_basal |
| rs12086549 | 1 | 119614337 | WARS2 | ENSG00000116874.7 | 6.126E-8 | 0 | 68957 | gtex_brain_putamen_basal |
| rs12081585 | 1 | 119615956 | WARS2 | ENSG00000116874.7 | 2.634E-9 | 0 | 67338 | gtex_brain_putamen_basal |
| rs7525871 | 1 | 119621894 | WARS2 | ENSG00000116874.7 | 2.183E-7 | 0 | 61400 | gtex_brain_putamen_basal |
| rs10802075 | 1 | 119627227 | WARS2 | ENSG00000116874.7 | 4.752E-9 | 0 | 56067 | gtex_brain_putamen_basal |
| rs61808897 | 1 | 119638200 | WARS2 | ENSG00000116874.7 | 2.586E-7 | 0 | 45094 | gtex_brain_putamen_basal |
| rs12064109 | 1 | 119642296 | WARS2 | ENSG00000116874.7 | 5.283E-7 | 0 | 40998 | gtex_brain_putamen_basal |
| rs61808932 | 1 | 119643820 | WARS2 | ENSG00000116874.7 | 1.02E-6 | 0 | 39474 | gtex_brain_putamen_basal |
| rs3936018 | 1 | 119682370 | WARS2 | ENSG00000116874.7 | 3.806E-9 | 0 | 924 | gtex_brain_putamen_basal |
| rs55696509 | 1 | 119686656 | WARS2 | ENSG00000116874.7 | 3.066E-9 | 0 | -3362 | gtex_brain_putamen_basal |
| rs72691108 | 1 | 119762175 | WARS2 | ENSG00000116874.7 | 8.97E-9 | 0 | -78881 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
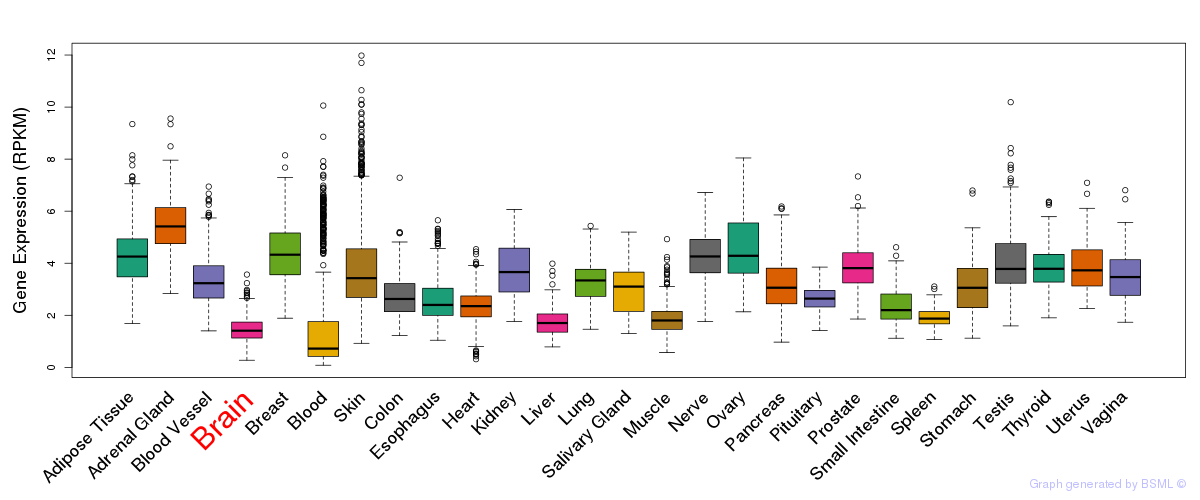
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NCOA3 | 0.94 | 0.96 |
| NR2C2 | 0.93 | 0.96 |
| CUGBP2 | 0.93 | 0.95 |
| AFF4 | 0.93 | 0.98 |
| RC3H2 | 0.93 | 0.97 |
| NBEA | 0.92 | 0.94 |
| MED13 | 0.92 | 0.97 |
| SPIRE1 | 0.92 | 0.93 |
| PUM2 | 0.92 | 0.96 |
| KIF1B | 0.91 | 0.97 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.65 | -0.81 |
| AF347015.31 | -0.65 | -0.81 |
| SERPINB6 | -0.65 | -0.70 |
| MT-CO2 | -0.64 | -0.81 |
| HIGD1B | -0.63 | -0.81 |
| HSD17B14 | -0.63 | -0.72 |
| AF347015.33 | -0.62 | -0.76 |
| IFI27 | -0.62 | -0.77 |
| S100B | -0.62 | -0.75 |
| S100A16 | -0.61 | -0.75 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004830 | tryptophan-tRNA ligase activity | IEA | - | |
| GO:0016874 | ligase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006436 | tryptophanyl-tRNA aminoacylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005759 | mitochondrial matrix | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TRYPTOPHAN METABOLISM | 40 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG AMINOACYL TRNA BIOSYNTHESIS | 41 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRNA AMINOACYLATION | 42 | 34 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| WONG MITOCHONDRIA GENE MODULE | 217 | 122 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |