Gene Page: SLU7
Summary ?
| GeneID | 10569 |
| Symbol | SLU7 |
| Synonyms | 9G8|hSlu7 |
| Description | SLU7 homolog, splicing factor |
| Reference | MIM:605974|HGNC:HGNC:16939|Ensembl:ENSG00000164609|HPRD:12074|Vega:OTTHUMG00000130324 |
| Gene type | protein-coding |
| Map location | 5q33.3 |
| Pascal p-value | 0.489 |
| Sherlock p-value | 0.466 |
| Fetal beta | -0.313 |
| DMG | 1 (# studies) |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01718 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00459 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02367472 | 5 | 159845898 | SLU7 | -0.022 | 0.48 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
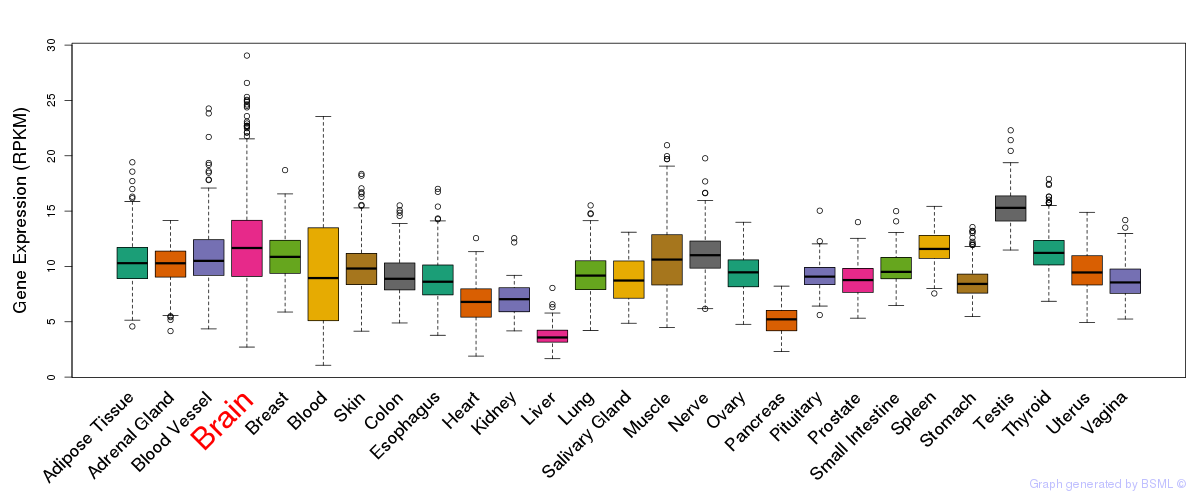
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000386 | second spliceosomal transesterification activity | IDA | 10197984 | |
| GO:0003676 | nucleic acid binding | IEA | - | |
| GO:0008270 | zinc ion binding | IDA | 15181151 | |
| GO:0030628 | pre-mRNA 3'-splice site binding | IDA | 15728250 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000380 | alternative nuclear mRNA splicing, via spliceosome | IDA | 15728250 | |
| GO:0000389 | nuclear mRNA 3'-splice site recognition | IDA | 15728250 | |
| GO:0006397 | mRNA processing | IEA | - | |
| GO:0008380 | RNA splicing | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005681 | spliceosome | IDA | 10197984 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016607 | nuclear speck | IDA | 15728250 | |
| GO:0030532 | small nuclear ribonucleoprotein complex | IDA | 15181151 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| MELLMAN TUT1 TARGETS UP | 19 | 11 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |