Gene Page: TIRAP
Summary ?
| GeneID | 114609 |
| Symbol | TIRAP |
| Synonyms | BACTS1|Mal|MyD88-2|wyatt |
| Description | toll-interleukin 1 receptor (TIR) domain containing adaptor protein |
| Reference | MIM:606252|HGNC:HGNC:17192|Ensembl:ENSG00000150455|Vega:OTTHUMG00000140373 |
| Gene type | protein-coding |
| Map location | 11q24.2 |
| Pascal p-value | 0.021 |
| Fetal beta | -0.937 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Cortex Frontal Cortex BA9 Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15932284 | 11 | 126153561 | TIRAP | 2.834E-4 | 0.432 | 0.039 | DMG:Wockner_2014 |
| cg13986503 | 11 | 126152983 | TIRAP | 3.951E-4 | -0.168 | 0.043 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs574198 | 11 | 126069841 | TIRAP | ENSG00000150455.9 | 1.849E-6 | 0.01 | -83119 | gtex_brain_ba24 |
| rs587891 | 11 | 126081336 | TIRAP | ENSG00000150455.9 | 1.602E-6 | 0.01 | -71624 | gtex_brain_ba24 |
| rs633607 | 11 | 126086768 | TIRAP | ENSG00000150455.9 | 1.602E-6 | 0.01 | -66192 | gtex_brain_ba24 |
| rs12293354 | 11 | 126093098 | TIRAP | ENSG00000150455.9 | 1.602E-6 | 0.01 | -59862 | gtex_brain_ba24 |
| rs10790794 | 11 | 126093217 | TIRAP | ENSG00000150455.9 | 1.602E-6 | 0.01 | -59743 | gtex_brain_ba24 |
| rs10893488 | 11 | 126095747 | TIRAP | ENSG00000150455.9 | 1.602E-6 | 0.01 | -57213 | gtex_brain_ba24 |
| rs675412 | 11 | 126103368 | TIRAP | ENSG00000150455.9 | 2.143E-7 | 0.01 | -49592 | gtex_brain_ba24 |
| rs1815820 | 11 | 126111520 | TIRAP | ENSG00000150455.9 | 1.602E-6 | 0.01 | -41440 | gtex_brain_ba24 |
| rs642137 | 11 | 126114630 | TIRAP | ENSG00000150455.9 | 1.602E-6 | 0.01 | -38330 | gtex_brain_ba24 |
| rs685409 | 11 | 126130232 | TIRAP | ENSG00000150455.9 | 1.602E-6 | 0.01 | -22728 | gtex_brain_ba24 |
| rs609634 | 11 | 126163124 | TIRAP | ENSG00000150455.9 | 1.501E-6 | 0.01 | 10164 | gtex_brain_ba24 |
| rs625413 | 11 | 126164349 | TIRAP | ENSG00000150455.9 | 6.669E-7 | 0.01 | 11389 | gtex_brain_ba24 |
| rs682991 | 11 | 126167696 | TIRAP | ENSG00000150455.9 | 6.5E-7 | 0.01 | 14736 | gtex_brain_ba24 |
| rs2962116 | 11 | 126169009 | TIRAP | ENSG00000150455.9 | 1.373E-6 | 0.01 | 16049 | gtex_brain_ba24 |
| rs603758 | 11 | 126203737 | TIRAP | ENSG00000150455.9 | 2.093E-6 | 0.01 | 50777 | gtex_brain_ba24 |
Section II. Transcriptome annotation
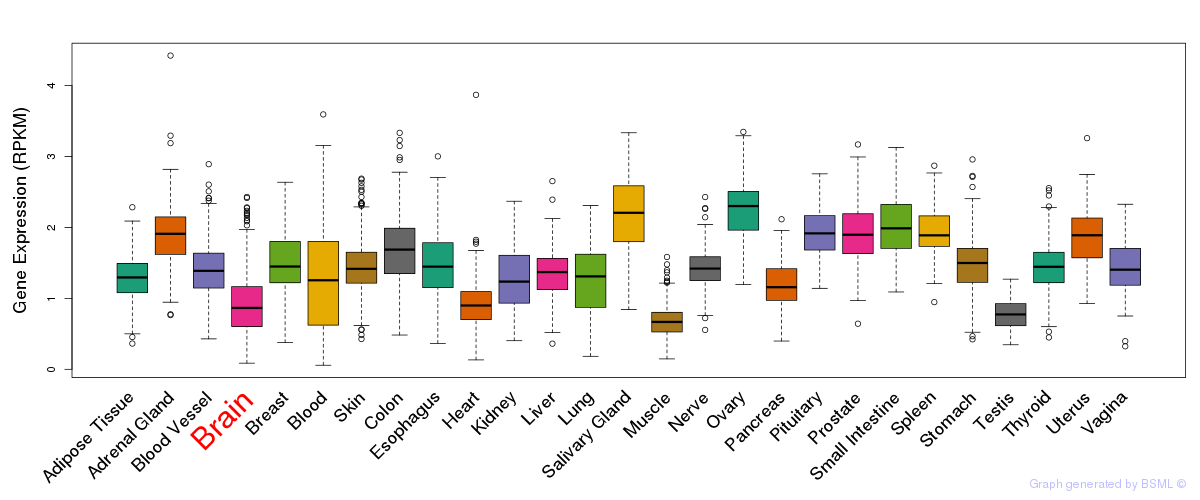
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GSK3 PATHWAY | 27 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TOLL PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| PID TOLL ENDOGENOUS PATHWAY | 25 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE DN | 77 | 49 | All SZGR 2.0 genes in this pathway |
| GOLDRATH IMMUNE MEMORY | 65 | 42 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| OUYANG PROSTATE CANCER PROGRESSION UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS DN | 145 | 93 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE UP | 36 | 24 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |