Gene Page: AP2M1
Summary ?
| GeneID | 1173 |
| Symbol | AP2M1 |
| Synonyms | AP50|CLAPM1|mu2 |
| Description | adaptor related protein complex 2 mu 1 subunit |
| Reference | MIM:601024|HGNC:HGNC:564|Ensembl:ENSG00000161203|HPRD:03014|Vega:OTTHUMG00000156822 |
| Gene type | protein-coding |
| Map location | 3q28 |
| Pascal p-value | 0.301 |
| Sherlock p-value | 0.826 |
| Fetal beta | -0.87 |
| DMG | 2 (# studies) |
| Support | ENDOCYTOSIS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17343451 | 3 | 183899704 | AP2M1 | 3.2E-4 | 0.006 | 0.184 | DMG:Montano_2016 |
| cg23679085 | 3 | 183892773 | AP2M1 | 7.35E-9 | -0.008 | 3.66E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
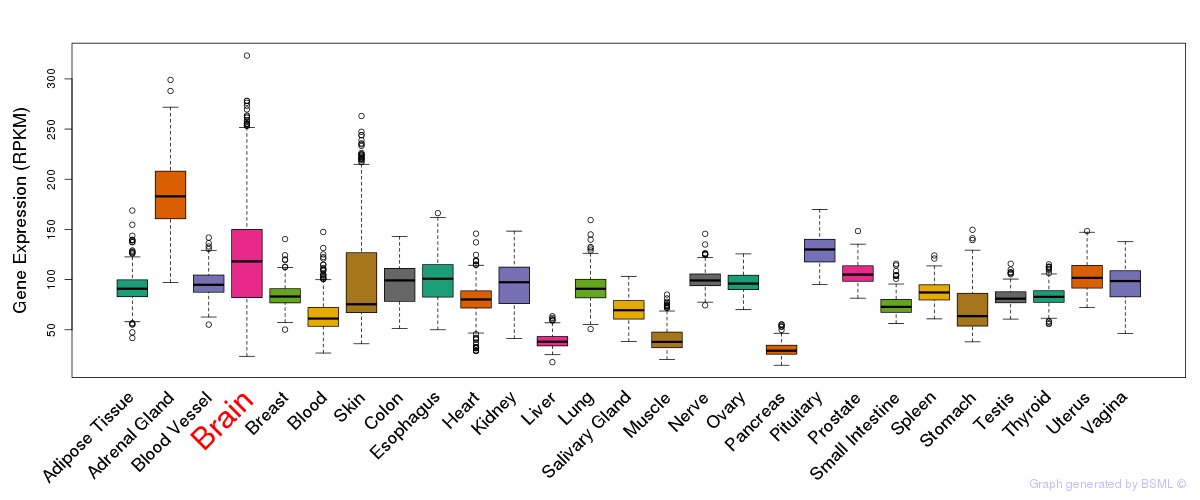
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TM9SF3 | 0.95 | 0.95 |
| FBXW11 | 0.92 | 0.91 |
| ZDHHC17 | 0.92 | 0.92 |
| LONP2 | 0.91 | 0.90 |
| PDZD8 | 0.91 | 0.91 |
| HIPK3 | 0.91 | 0.92 |
| GARNL1 | 0.90 | 0.91 |
| APPL1 | 0.90 | 0.92 |
| FAM91A1 | 0.90 | 0.92 |
| SERINC1 | 0.90 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.65 | -0.55 |
| FXYD1 | -0.62 | -0.55 |
| C1orf54 | -0.61 | -0.58 |
| MT-CO2 | -0.61 | -0.56 |
| HIGD1B | -0.60 | -0.55 |
| ENHO | -0.60 | -0.60 |
| IL32 | -0.60 | -0.52 |
| VAMP5 | -0.59 | -0.49 |
| CSAG1 | -0.59 | -0.53 |
| C1orf61 | -0.58 | -0.63 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADRA1B | ADRA1 | ALPHA1BAR | adrenergic, alpha-1B-, receptor | Affinity Capture-Western Two-hybrid | BioGRID | 12644451 |
| AP2B1 | ADTB2 | AP105B | AP2-BETA | CLAPB1 | DKFZp781K0743 | adaptor-related protein complex 2, beta 1 subunit | Two-hybrid | BioGRID | 16189514 |
| AQP4 | HMIWC2 | MGC22454 | MIWC | aquaporin 4 | - | HPRD,BioGRID | 11742978 |
| CD22 | FLJ22814 | MGC130020 | SIGLEC-2 | SIGLEC2 | CD22 molecule | - | HPRD,BioGRID | 12646615 |
| CLEC4M | CD209L | CD299 | DC-SIGN2 | DC-SIGNR | DCSIGNR | HP10347 | L-SIGN | LSIGN | MGC129964 | MGC47866 | C-type lectin domain family 4, member M | Affinity Capture-Western | BioGRID | 12621057 |
| CTLA4 | CD152 | CELIAC3 | CTLA-4 | GSE | IDDM12 | cytotoxic T-lymphocyte-associated protein 4 | Affinity Capture-Western Co-crystal Structure Reconstituted Complex Two-hybrid | BioGRID | 9200449 |11583591 |
| CTLA4 | CD152 | CELIAC3 | CTLA-4 | GSE | IDDM12 | cytotoxic T-lymphocyte-associated protein 4 | - | HPRD | 9200449 |9256472 |11583591|9256472 |
| DAB2 | DOC-2 | DOC2 | FLJ26626 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | - | HPRD | 11247302 |
| DCX | DBCN | DC | LISX | SCLH | XLIS | doublecortin | - | HPRD | 11591131 |
| DPPA2 | PESCRG1 | developmental pluripotency associated 2 | Two-hybrid | BioGRID | 16189514 |
| DVL2 | - | dishevelled, dsh homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| EHD2 | FLJ96617 | PAST2 | EH-domain containing 2 | - | HPRD,BioGRID | 15182197 |
| FURIN | FUR | PACE | PCSK3 | SPC1 | furin (paired basic amino acid cleaving enzyme) | - | HPRD,BioGRID | 10593987 |
| FXR2 | FMR1L2 | fragile X mental retardation, autosomal homolog 2 | Two-hybrid | BioGRID | 16189514 |
| IKZF1 | Hs.54452 | IK1 | IKAROS | LYF1 | PRO0758 | ZNFN1A1 | hIk-1 | IKAROS family zinc finger 1 (Ikaros) | Two-hybrid | BioGRID | 16189514 |
| L1CAM | CAML1 | CD171 | HSAS | HSAS1 | MASA | MIC5 | N-CAML1 | S10 | SPG1 | L1 cell adhesion molecule | L1CAM interacts with AP-2 mu2 chain. This interaction was modelled on a demonstrated interaction between human L1CAM and AP-2 mu2 chain from an unspecified species. | BIND | 15647482 |
| LAMP1 | CD107a | LAMPA | LGP120 | lysosomal-associated membrane protein 1 | - | HPRD,BioGRID | 7569928 |
| LY9 | CD229 | SLAMF3 | hly9 | mLY9 | lymphocyte antigen 9 | - | HPRD,BioGRID | 12621057 |
| MED4 | DRIP36 | FLJ10956 | HSPC126 | RP11-90M2.2 | TRAP36 | VDRIP | mediator complex subunit 4 | Two-hybrid | BioGRID | 16189514 |
| MEGF10 | DKFZp781K1852 | FLJ41574 | KIAA1780 | multiple EGF-like-domains 10 | - | HPRD | 12421765 |
| RALBP1 | RIP1 | RLIP1 | RLIP76 | ralA binding protein 1 | RLIP76 interacts with mu-2. | BIND | 10910768 |
| RTDR1 | MGC16968 | rhabdoid tumor deletion region gene 1 | Two-hybrid | BioGRID | 16189514 |
| RUNDC3A | RAP2IP | RPIP8 | RUN domain containing 3A | Two-hybrid | BioGRID | 16189514 |
| STON2 | STN2 | STNB | STNB2 | stonin 2 | Reconstituted Complex | BioGRID | 11454741 |
| TBC1D5 | KIAA0210 | TBC1 domain family, member 5 | Two-hybrid | BioGRID | 16189514 |
| TGOLN2 | MGC14722 | TGN38 | TGN46 | TGN48 | TGN51 | TTGN2 | trans-golgi network protein 2 | TGN38 interacts with Mu2. This interaction was modeled on a demonstrated interaction between TGN38 from unspecified species and rat Mu2. | BIND | 15916959 |
| ZBTB8 | BOZF1 | FLJ90065 | MGC17919 | zinc finger and BTB domain containing 8 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NDKDYNAMIN PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PTDINS PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BARRESTIN PATHWAY | 10 | 8 | All SZGR 2.0 genes in this pathway |
| PID ARF 3PATHWAY | 19 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME GAP JUNCTION DEGRADATION | 10 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME MEMBRANE TRAFFICKING | 129 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | 21 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME MHC CLASS II ANTIGEN PRESENTATION | 91 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | 13 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME EGFR DOWNREGULATION | 25 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAFFICKING OF AMPA RECEPTORS | 28 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | 16 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME L1CAM INTERACTIONS | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNAL TRANSDUCTION BY L1 | 34 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME RECYCLING PATHWAY OF L1 | 27 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | 28 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME GAP JUNCTION TRAFFICKING | 27 | 19 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS UP | 112 | 68 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| RAMPON ENRICHED LEARNING ENVIRONMENT EARLY UP | 15 | 12 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX DN | 54 | 43 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |