Gene Page: CLC
Summary ?
| GeneID | 1178 |
| Symbol | CLC |
| Synonyms | GAL10|Gal-10|LGALS10|LGALS10A|LPPL_HUMAN |
| Description | Charcot-Leyden crystal galectin |
| Reference | MIM:153310|HGNC:HGNC:2014|Ensembl:ENSG00000105205|HPRD:08351|Vega:OTTHUMG00000183144 |
| Gene type | protein-coding |
| Map location | 19q13.1 |
| Pascal p-value | 0.294 |
| Fetal beta | 0.963 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
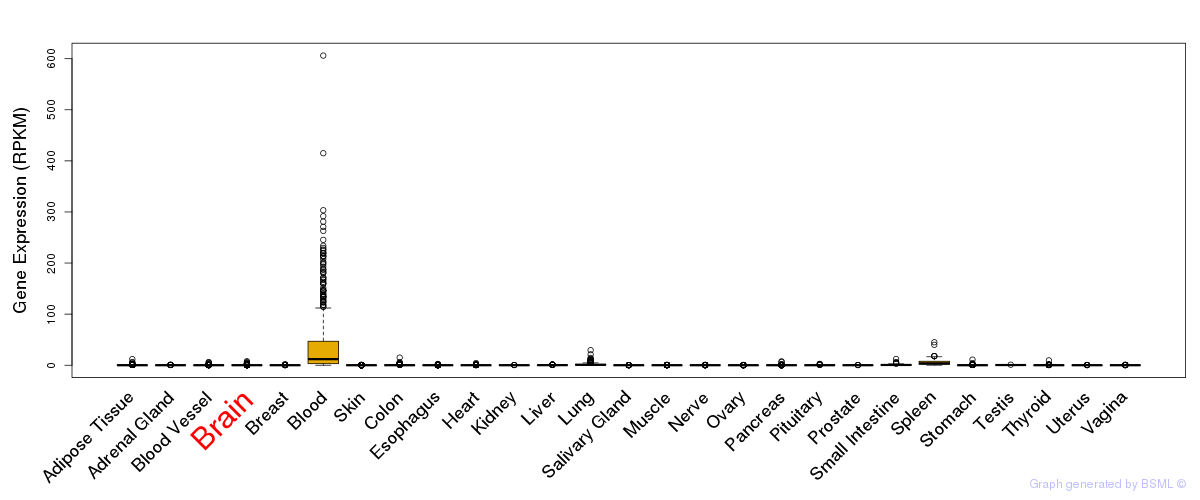
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TRIM7 | 0.41 | 0.29 |
| FUT7 | 0.40 | 0.31 |
| SLC22A20 | 0.39 | 0.28 |
| AL133215.4 | 0.38 | 0.30 |
| BEST4 | 0.38 | 0.37 |
| C15orf62 | 0.37 | 0.30 |
| MAPK12 | 0.37 | 0.26 |
| INSRR | 0.37 | 0.27 |
| TFAP2E | 0.37 | 0.33 |
| C3orf62 | 0.36 | 0.29 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SCG3 | -0.23 | -0.24 |
| LYRM5 | -0.20 | -0.23 |
| IFIH1 | -0.20 | -0.22 |
| CBR3 | -0.19 | -0.21 |
| FAM162A | -0.19 | -0.18 |
| RLBP1 | -0.18 | -0.20 |
| FAM162B | -0.18 | -0.17 |
| PRDX1 | -0.18 | -0.16 |
| ELTD1 | -0.18 | -0.19 |
| SEPT7 | -0.17 | -0.16 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINED IN MONOCYTE DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS CML DIVIDING DN | 11 | 8 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL DIVIDING UP | 13 | 7 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT UP | 181 | 101 | All SZGR 2.0 genes in this pathway |
| WANG METHYLATED IN BREAST CANCER | 35 | 25 | All SZGR 2.0 genes in this pathway |
| NAKAJIMA EOSINOPHIL | 30 | 20 | All SZGR 2.0 genes in this pathway |
| KUUSELO PANCREATIC CANCER 19Q13 AMPLIFICATION | 35 | 22 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION C | 69 | 49 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM HIGH RISK DN | 20 | 13 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION DN | 99 | 65 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ WALDENSTROEMS MACROGLOBULINEMIA 2 | 13 | 9 | All SZGR 2.0 genes in this pathway |
| GENTLES LEUKEMIC STEM CELL DN | 19 | 10 | All SZGR 2.0 genes in this pathway |
| NABA ECM AFFILIATED | 171 | 89 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |