Gene Page: CPNE4
Summary ?
| GeneID | 131034 |
| Symbol | CPNE4 |
| Synonyms | COPN4|CPN4 |
| Description | copine 4 |
| Reference | MIM:604208|HGNC:HGNC:2317|Ensembl:ENSG00000196353|HPRD:05017| |
| Gene type | protein-coding |
| Map location | 3q22.1 |
| Pascal p-value | 0.019 |
| Sherlock p-value | 0.97 |
| Fetal beta | -2.183 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | STRUCTURAL PLASTICITY |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18512963 | 3 | 131430364 | CPNE4 | 4.565E-4 | -0.453 | 0.046 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10969802 | chr9 | 30636427 | CPNE4 | 131034 | 0.09 | trans | ||
| rs10969816 | chr9 | 30661611 | CPNE4 | 131034 | 0.09 | trans | ||
| rs12350524 | chr9 | 30743421 | CPNE4 | 131034 | 0.09 | trans | ||
| rs12343068 | chr9 | 30783524 | CPNE4 | 131034 | 0.09 | trans | ||
| rs10511868 | chr9 | 30816504 | CPNE4 | 131034 | 0.09 | trans |
Section II. Transcriptome annotation
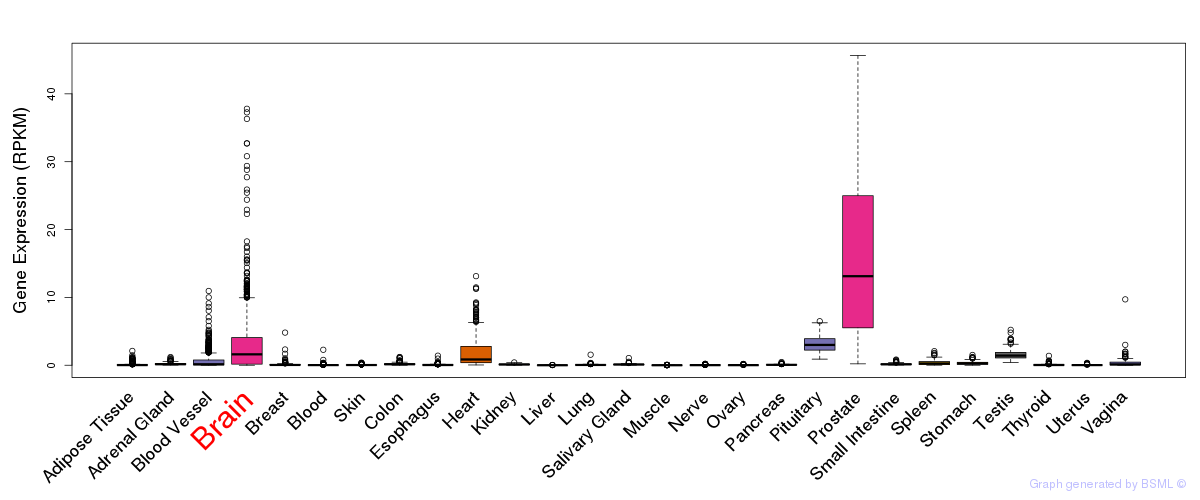
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTB | PS1TP5BP1 | actin, beta | - | HPRD | 12522145 |
| BCOR | ANOP2 | FLJ20285 | FLJ38041 | KIAA1575 | MAA2 | MCOPS2 | MGC131961 | MGC71031 | BCL6 co-repressor | - | HPRD | 12522145 |
| CCDC22 | CXorf37 | JM1 | coiled-coil domain containing 22 | - | HPRD | 12522145 |
| CPNE1 | COPN1 | CPN1 | MGC1142 | copine I | - | HPRD | 12522145 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | - | HPRD | 12522145 |
| MYCBP | AMY-1 | c-myc binding protein | - | HPRD | 12522145 |
| PDCD6 | ALG-2 | FLJ46208 | MGC111017 | MGC119050 | MGC9123 | PEF1B | programmed cell death 6 | - | HPRD | 12522145 |
| PITPNM2 | KIAA1457 | NIR3 | RDGB2 | RDGBA2 | phosphatidylinositol transfer protein, membrane-associated 2 | - | HPRD | 12522145 |
| PPP5C | FLJ36922 | PP5 | PPP5 | protein phosphatase 5, catalytic subunit | - | HPRD | 12522145 |
| RDX | DFNB24 | radixin | - | HPRD | 12522145 |
| SKIL | SNO | SnoA | SnoN | SKI-like oncogene | - | HPRD | 12522145 |
| SPTBN1 | ELF | SPTB2 | betaSpII | spectrin, beta, non-erythrocytic 1 | - | HPRD | 12522145 |
| UBE2M | UBC-RS2 | UBC12 | hUbc12 | ubiquitin-conjugating enzyme E2M (UBC12 homolog, yeast) | - | HPRD | 12522145 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS UP | 43 | 28 | All SZGR 2.0 genes in this pathway |