Gene Page: GNPDA2
Summary ?
| GeneID | 132789 |
| Symbol | GNPDA2 |
| Synonyms | GNP2|SB52 |
| Description | glucosamine-6-phosphate deaminase 2 |
| Reference | MIM:613222|HGNC:HGNC:21526|Ensembl:ENSG00000163281|HPRD:13594|Vega:OTTHUMG00000099415 |
| Gene type | protein-coding |
| Map location | 4p12 |
| Pascal p-value | 0.238 |
| Sherlock p-value | 0.649 |
| DEG p-value | DEG:Sanders_2014:DS1_p=0.132:DS1_beta=0.039200:DS2_p=5.12e-01:DS2_beta=0.030:DS2_FDR=7.35e-01 |
| Fetal beta | -0.307 |
| eGene | Caudate basal ganglia Cerebellum Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Sanders_2013 | Microarray | Whole-genome gene expression profiles using microarrays on lymphoblastoid cell lines (LCLs) from 413 cases and 446 controls. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16857236 | 4 | 44614810 | GNPDA2 | ENSG00000163281.7 | 4.07E-7 | 0 | 113802 | gtex_brain_putamen_basal |
| rs11736925 | 4 | 44616283 | GNPDA2 | ENSG00000163281.7 | 1.813E-8 | 0 | 112329 | gtex_brain_putamen_basal |
| rs11734057 | 4 | 44616296 | GNPDA2 | ENSG00000163281.7 | 1.813E-8 | 0 | 112316 | gtex_brain_putamen_basal |
| rs17600797 | 4 | 44626806 | GNPDA2 | ENSG00000163281.7 | 8.681E-8 | 0 | 101806 | gtex_brain_putamen_basal |
| rs6831166 | 4 | 44630977 | GNPDA2 | ENSG00000163281.7 | 1.197E-7 | 0 | 97635 | gtex_brain_putamen_basal |
| rs6850107 | 4 | 44633866 | GNPDA2 | ENSG00000163281.7 | 1.353E-7 | 0 | 94746 | gtex_brain_putamen_basal |
| rs6813944 | 4 | 44635157 | GNPDA2 | ENSG00000163281.7 | 1.352E-7 | 0 | 93455 | gtex_brain_putamen_basal |
| rs10938361 | 4 | 44638931 | GNPDA2 | ENSG00000163281.7 | 1.014E-9 | 0 | 89681 | gtex_brain_putamen_basal |
| rs7679737 | 4 | 44639517 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 89095 | gtex_brain_putamen_basal |
| rs4235132 | 4 | 44641392 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 87220 | gtex_brain_putamen_basal |
| rs34018268 | 4 | 44644463 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 84149 | gtex_brain_putamen_basal |
| rs12646633 | 4 | 44644687 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 83925 | gtex_brain_putamen_basal |
| rs6852372 | 4 | 44651820 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 76792 | gtex_brain_putamen_basal |
| rs1388310 | 4 | 44653414 | GNPDA2 | ENSG00000163281.7 | 1.497E-8 | 0 | 75198 | gtex_brain_putamen_basal |
| rs369684976 | 4 | 44657868 | GNPDA2 | ENSG00000163281.7 | 1.181E-8 | 0 | 70744 | gtex_brain_putamen_basal |
| rs12639722 | 4 | 44660248 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 68364 | gtex_brain_putamen_basal |
| rs62305366 | 4 | 44664907 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 63705 | gtex_brain_putamen_basal |
| rs35372782 | 4 | 44667338 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 61274 | gtex_brain_putamen_basal |
| rs62305368 | 4 | 44673826 | GNPDA2 | ENSG00000163281.7 | 9.515E-10 | 0 | 54786 | gtex_brain_putamen_basal |
| rs200576312 | 4 | 44677677 | GNPDA2 | ENSG00000163281.7 | 3.82E-8 | 0 | 50935 | gtex_brain_putamen_basal |
| rs75324498 | 4 | 44677678 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 50934 | gtex_brain_putamen_basal |
| rs10461070 | 4 | 44678737 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 49875 | gtex_brain_putamen_basal |
| rs11734296 | 4 | 44679560 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 49052 | gtex_brain_putamen_basal |
| rs62305372 | 4 | 44683541 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 45071 | gtex_brain_putamen_basal |
| rs11931747 | 4 | 44687163 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 41449 | gtex_brain_putamen_basal |
| rs12499960 | 4 | 44687401 | GNPDA2 | ENSG00000163281.7 | 6.811E-8 | 0 | 41211 | gtex_brain_putamen_basal |
| rs12639842 | 4 | 44691098 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 37514 | gtex_brain_putamen_basal |
| rs71611910 | 4 | 44693499 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 35113 | gtex_brain_putamen_basal |
| rs2306992 | 4 | 44694077 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 34535 | gtex_brain_putamen_basal |
| rs201814154 | 4 | 44695520 | GNPDA2 | ENSG00000163281.7 | 9.669E-9 | 0 | 33092 | gtex_brain_putamen_basal |
| rs77297867 | 4 | 44695521 | GNPDA2 | ENSG00000163281.7 | 3.36E-9 | 0 | 33091 | gtex_brain_putamen_basal |
| rs13125773 | 4 | 44701256 | GNPDA2 | ENSG00000163281.7 | 1.141E-8 | 0 | 27356 | gtex_brain_putamen_basal |
| rs1565532 | 4 | 44702720 | GNPDA2 | ENSG00000163281.7 | 9.534E-10 | 0 | 25892 | gtex_brain_putamen_basal |
| rs16857402 | 4 | 44706453 | GNPDA2 | ENSG00000163281.7 | 9.859E-10 | 0 | 22159 | gtex_brain_putamen_basal |
| rs2709 | 4 | 44706919 | GNPDA2 | ENSG00000163281.7 | 1.005E-9 | 0 | 21693 | gtex_brain_putamen_basal |
| rs62305383 | 4 | 44708086 | GNPDA2 | ENSG00000163281.7 | 9.534E-10 | 0 | 20526 | gtex_brain_putamen_basal |
| rs71648801 | 4 | 44723646 | GNPDA2 | ENSG00000163281.7 | 1.564E-9 | 0 | 4966 | gtex_brain_putamen_basal |
| rs11945975 | 4 | 44724566 | GNPDA2 | ENSG00000163281.7 | 1.185E-8 | 0 | 4046 | gtex_brain_putamen_basal |
| rs11724396 | 4 | 44725334 | GNPDA2 | ENSG00000163281.7 | 1.003E-9 | 0 | 3278 | gtex_brain_putamen_basal |
| rs16857442 | 4 | 44739911 | GNPDA2 | ENSG00000163281.7 | 1.637E-7 | 0 | -11299 | gtex_brain_putamen_basal |
| rs1491300 | 4 | 44742727 | GNPDA2 | ENSG00000163281.7 | 1.652E-7 | 0 | -14115 | gtex_brain_putamen_basal |
| rs138839217 | 4 | 44747451 | GNPDA2 | ENSG00000163281.7 | 2.135E-7 | 0 | -18839 | gtex_brain_putamen_basal |
| rs112680663 | 4 | 44749297 | GNPDA2 | ENSG00000163281.7 | 8.176E-7 | 0 | -20685 | gtex_brain_putamen_basal |
| rs13112967 | 4 | 44750803 | GNPDA2 | ENSG00000163281.7 | 2.198E-7 | 0 | -22191 | gtex_brain_putamen_basal |
| rs11934131 | 4 | 44761571 | GNPDA2 | ENSG00000163281.7 | 8.176E-7 | 0 | -32959 | gtex_brain_putamen_basal |
| rs146316923 | 4 | 44762368 | GNPDA2 | ENSG00000163281.7 | 7.248E-7 | 0 | -33756 | gtex_brain_putamen_basal |
| rs7675035 | 4 | 44764783 | GNPDA2 | ENSG00000163281.7 | 8.176E-7 | 0 | -36171 | gtex_brain_putamen_basal |
| rs12644039 | 4 | 44766644 | GNPDA2 | ENSG00000163281.7 | 3.348E-8 | 0 | -38032 | gtex_brain_putamen_basal |
| rs35152931 | 4 | 44766932 | GNPDA2 | ENSG00000163281.7 | 8.176E-7 | 0 | -38320 | gtex_brain_putamen_basal |
| rs11729477 | 4 | 44770333 | GNPDA2 | ENSG00000163281.7 | 7.836E-7 | 0 | -41721 | gtex_brain_putamen_basal |
| rs16857479 | 4 | 44770720 | GNPDA2 | ENSG00000163281.7 | 8.176E-7 | 0 | -42108 | gtex_brain_putamen_basal |
| rs11724366 | 4 | 44785512 | GNPDA2 | ENSG00000163281.7 | 2.171E-7 | 0 | -56900 | gtex_brain_putamen_basal |
| rs6816989 | 4 | 44791911 | GNPDA2 | ENSG00000163281.7 | 8.147E-7 | 0 | -63299 | gtex_brain_putamen_basal |
| rs71984973 | 4 | 44793288 | GNPDA2 | ENSG00000163281.7 | 7.891E-7 | 0 | -64676 | gtex_brain_putamen_basal |
| rs11728530 | 4 | 44794111 | GNPDA2 | ENSG00000163281.7 | 8.08E-7 | 0 | -65499 | gtex_brain_putamen_basal |
| rs36047545 | 4 | 44795072 | GNPDA2 | ENSG00000163281.7 | 1.47E-6 | 0 | -66460 | gtex_brain_putamen_basal |
| rs7683933 | 4 | 44798080 | GNPDA2 | ENSG00000163281.7 | 8.05E-7 | 0 | -69468 | gtex_brain_putamen_basal |
| rs12639624 | 4 | 44801988 | GNPDA2 | ENSG00000163281.7 | 8.052E-7 | 0 | -73376 | gtex_brain_putamen_basal |
| rs57068467 | 4 | 44806404 | GNPDA2 | ENSG00000163281.7 | 1.67E-7 | 0 | -77792 | gtex_brain_putamen_basal |
| rs12639682 | 4 | 44811032 | GNPDA2 | ENSG00000163281.7 | 1.67E-7 | 0 | -82420 | gtex_brain_putamen_basal |
| rs36022022 | 4 | 44818450 | GNPDA2 | ENSG00000163281.7 | 1.67E-7 | 0 | -89838 | gtex_brain_putamen_basal |
| rs11730341 | 4 | 44823800 | GNPDA2 | ENSG00000163281.7 | 4.348E-7 | 0 | -95188 | gtex_brain_putamen_basal |
| rs34521789 | 4 | 44824918 | GNPDA2 | ENSG00000163281.7 | 1.67E-7 | 0 | -96306 | gtex_brain_putamen_basal |
| rs62409354 | 4 | 44825505 | GNPDA2 | ENSG00000163281.7 | 7.065E-8 | 0 | -96893 | gtex_brain_putamen_basal |
| rs35765022 | 4 | 44827368 | GNPDA2 | ENSG00000163281.7 | 3.575E-8 | 0 | -98756 | gtex_brain_putamen_basal |
| rs11734975 | 4 | 44827537 | GNPDA2 | ENSG00000163281.7 | 1.757E-7 | 0 | -98925 | gtex_brain_putamen_basal |
| rs71816407 | 4 | 44832654 | GNPDA2 | ENSG00000163281.7 | 1.852E-7 | 0 | -104042 | gtex_brain_putamen_basal |
| rs12642145 | 4 | 44834738 | GNPDA2 | ENSG00000163281.7 | 1.739E-7 | 0 | -106126 | gtex_brain_putamen_basal |
| rs143320673 | 4 | 44835036 | GNPDA2 | ENSG00000163281.7 | 1.922E-7 | 0 | -106424 | gtex_brain_putamen_basal |
| rs924176 | 4 | 44842838 | GNPDA2 | ENSG00000163281.7 | 1.682E-7 | 0 | -114226 | gtex_brain_putamen_basal |
| rs4145971 | 4 | 44843214 | GNPDA2 | ENSG00000163281.7 | 1.682E-7 | 0 | -114602 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
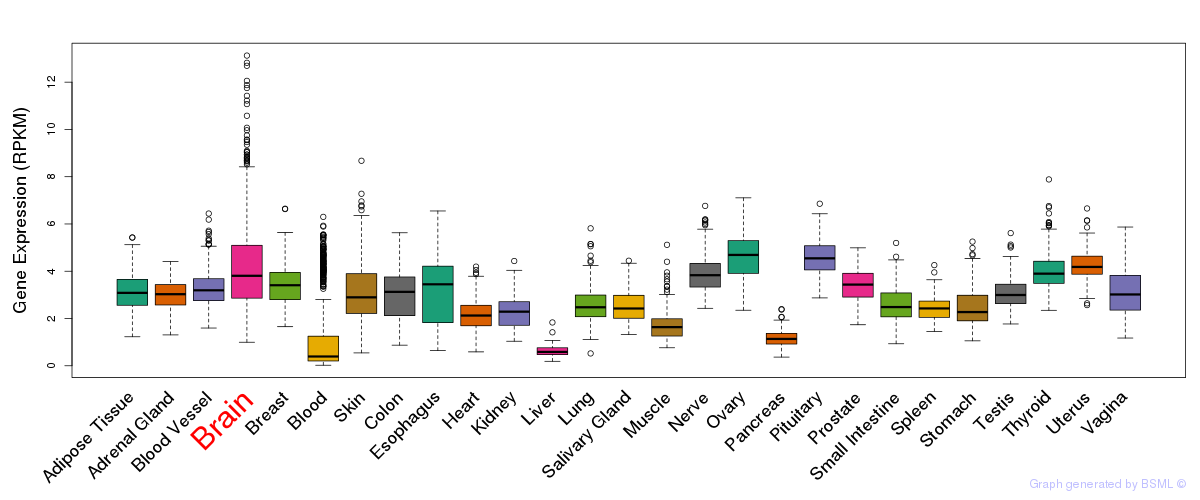
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AMINO SUGAR AND NUCLEOTIDE SUGAR METABOLISM | 44 | 30 | All SZGR 2.0 genes in this pathway |
| MAYBURD RESPONSE TO L663536 DN | 56 | 32 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS | 206 | 127 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR1 TARGETS UP | 53 | 39 | All SZGR 2.0 genes in this pathway |