Gene Page: CPN1
Summary ?
| GeneID | 1369 |
| Symbol | CPN1 |
| Synonyms | CPN|SCPN |
| Description | carboxypeptidase N subunit 1 |
| Reference | MIM:603103|HGNC:HGNC:2312|Ensembl:ENSG00000120054|HPRD:09120|Vega:OTTHUMG00000018896 |
| Gene type | protein-coding |
| Map location | 10q24.2 |
| Pascal p-value | 0.73 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15457079 | 10 | 101841816 | CPN1 | 3.326E-4 | -0.478 | 0.041 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
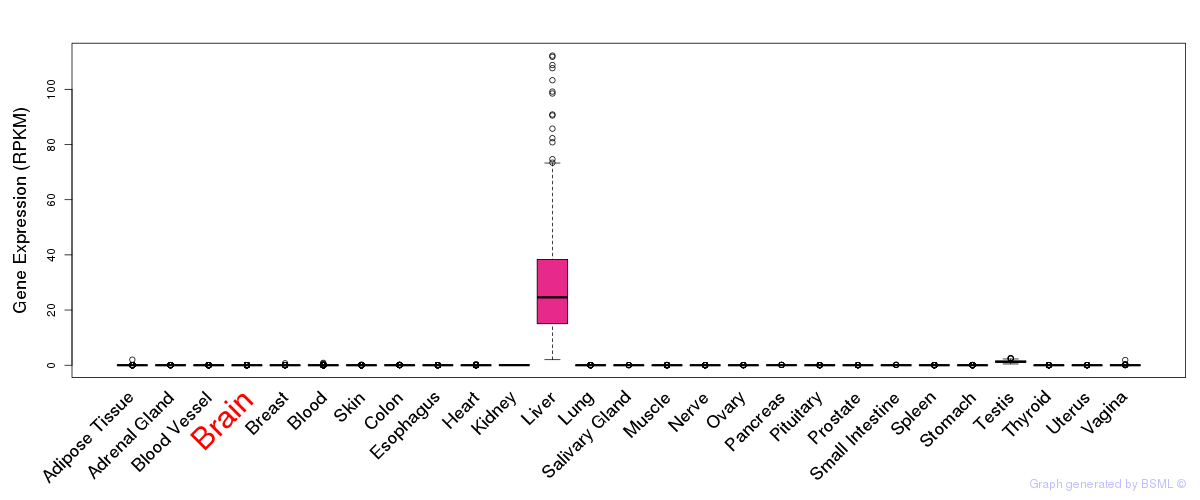
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SELL | 0.46 | 0.37 |
| HAVCR2 | 0.44 | 0.44 |
| LIPA | 0.42 | 0.45 |
| FECH | 0.42 | 0.49 |
| C3AR1 | 0.42 | 0.44 |
| TMEM64 | 0.42 | 0.43 |
| EVI2B | 0.42 | 0.37 |
| EDIL3 | 0.41 | 0.45 |
| MAN1A1 | 0.41 | 0.41 |
| RHOU | 0.40 | 0.40 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RP9P | -0.23 | -0.27 |
| IL32 | -0.22 | -0.17 |
| AF347015.18 | -0.22 | -0.23 |
| NSBP1 | -0.22 | -0.24 |
| AC098691.2 | -0.21 | -0.23 |
| AC005921.3 | -0.21 | -0.23 |
| AC010300.1 | -0.21 | -0.29 |
| AL022328.1 | -0.21 | -0.17 |
| AC007216.4 | -0.20 | -0.16 |
| AC022692.2 | -0.20 | -0.20 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE DN | 165 | 104 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS DN | 61 | 47 | All SZGR 2.0 genes in this pathway |
| SU LIVER | 55 | 32 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D UP | 89 | 62 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE DN | 138 | 99 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL DN | 113 | 76 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| KAMMINGA SENESCENCE | 41 | 26 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS DN | 109 | 71 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS UP | 266 | 142 | All SZGR 2.0 genes in this pathway |