Gene Page: CRYAA
Summary ?
| GeneID | 1409 |
| Symbol | CRYAA |
| Synonyms | CRYA1|CTRCT9|HSPB4 |
| Description | crystallin alpha A |
| Reference | MIM:123580|HGNC:HGNC:2388|Ensembl:ENSG00000160202|HPRD:00427|Vega:OTTHUMG00000086842 |
| Gene type | protein-coding |
| Map location | 21q22.3 |
| Pascal p-value | 0.038 |
| Fetal beta | -0.695 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
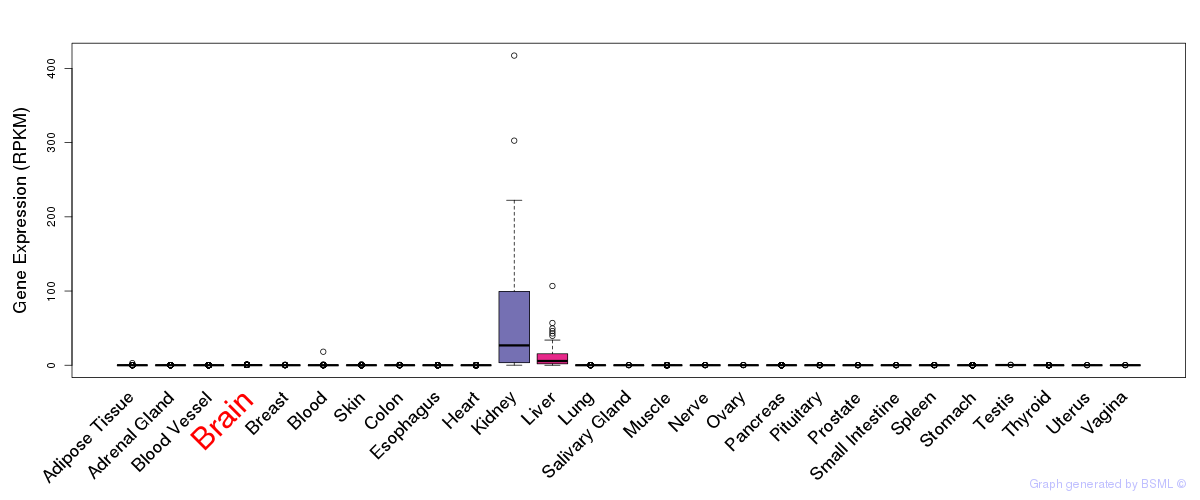
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C16orf79 | 0.44 | 0.33 |
| NSUN5C | 0.41 | 0.35 |
| AL022328.1 | 0.41 | 0.33 |
| NSUN5B | 0.40 | 0.32 |
| AC011511.1 | 0.40 | 0.32 |
| NEIL1 | 0.40 | 0.34 |
| GSDMB | 0.38 | 0.33 |
| FGF17 | 0.38 | 0.26 |
| ZNF692 | 0.38 | 0.29 |
| CCDC84 | 0.37 | 0.29 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KLHL5 | -0.30 | -0.29 |
| ARHGAP5 | -0.30 | -0.27 |
| NET1 | -0.30 | -0.26 |
| TMED7 | -0.30 | -0.27 |
| PDE3A | -0.30 | -0.28 |
| ABCD2 | -0.29 | -0.25 |
| TNFRSF1B | -0.29 | -0.24 |
| TICAM2 | -0.29 | -0.26 |
| TBC1D12 | -0.29 | -0.27 |
| VWF | -0.29 | -0.24 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ALB | DKFZp779N1935 | PRO0883 | PRO0903 | PRO1341 | albumin | - | HPRD | 9618718 |
| BAX | BCL2L4 | BCL2-associated X protein | HalphaA-crystallin interacts with Bax. | BIND | 14752512 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | HalphaA-crystallin interacts with Bcl-Xs. | BIND | 14752512 |
| CRYAA | CRYA1 | HSPB4 | crystallin, alpha A | Two-hybrid | BioGRID | 11700327 |
| CRYAB | CRYA2 | CTPP2 | HSPB5 | crystallin, alpha B | Affinity Capture-Western Two-hybrid | BioGRID | 11700327 |
| CRYBB2 | CCA2 | CRYB2 | CRYB2A | D22S665 | crystallin, beta B2 | Affinity Capture-Western Two-hybrid | BioGRID | 11700327 |
| CRYGC | CCL | CRYG3 | crystallin, gamma C | Affinity Capture-Western Two-hybrid | BioGRID | 11700327 |
| CRYZ | DKFZp779E0834 | FLJ41475 | crystallin, zeta (quinone reductase) | - | HPRD | 11672428 |
| CS | - | citrate synthase | - | HPRD | 11377425 |
| HSPB1 | CMT2F | DKFZp586P1322 | HMN2B | HS.76067 | HSP27 | HSP28 | Hsp25 | SRP27 | heat shock 27kDa protein 1 | - | HPRD,BioGRID | 11700327 |
| LALBA | MGC138521 | MGC138523 | lactalbumin, alpha- | - | HPRD,BioGRID | 9346914 |
| MIP | AQP0 | LIM1 | MIP26 | MP26 | major intrinsic protein of lens fiber | - | HPRD,BioGRID | 8910261 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE DN | 87 | 59 | All SZGR 2.0 genes in this pathway |
| LIU SMARCA4 TARGETS | 64 | 39 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 UP | 107 | 67 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE UP | 36 | 24 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS POLYSOMY7 UP | 79 | 50 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K4ME3 AND H327ME3 | 126 | 83 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |