Gene Page: DAZL
Summary ?
| GeneID | 1618 |
| Symbol | DAZL |
| Synonyms | DAZH|DAZL1|DAZLA|SPGYLA |
| Description | deleted in azoospermia like |
| Reference | MIM:601486|HGNC:HGNC:2685|Ensembl:ENSG00000092345|HPRD:09029|Vega:OTTHUMG00000157050 |
| Gene type | protein-coding |
| Map location | 3p24.3 |
| Pascal p-value | 0.358 |
| Fetal beta | -0.348 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16639713 | 3 | 16646904 | DAZL | 5.651E-4 | 0.368 | 0.049 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16869851 | chr4 | 20690056 | DAZL | 1618 | 0.12 | trans |
Section II. Transcriptome annotation
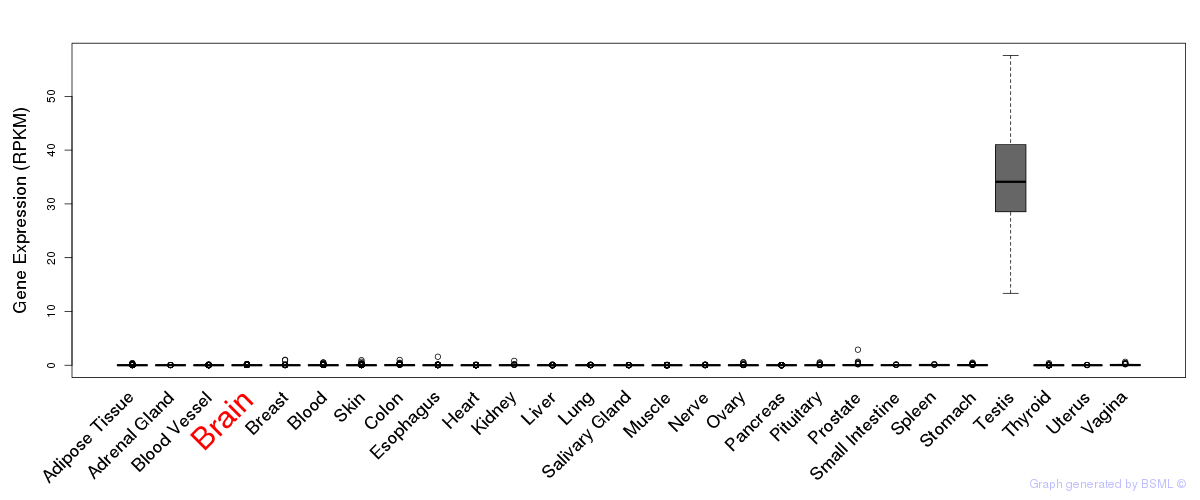
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NFIA | 0.87 | 0.77 |
| IGSF3 | 0.85 | 0.91 |
| NFIB | 0.85 | 0.86 |
| XPR1 | 0.85 | 0.92 |
| BCL11A | 0.84 | 0.85 |
| ITSN1 | 0.84 | 0.84 |
| EPHB1 | 0.84 | 0.81 |
| NFYA | 0.84 | 0.90 |
| VANGL2 | 0.84 | 0.81 |
| C10orf12 | 0.84 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TNFSF12 | -0.52 | -0.64 |
| C5orf53 | -0.52 | -0.76 |
| TMEM54 | -0.51 | -0.56 |
| PTH1R | -0.50 | -0.72 |
| LHPP | -0.50 | -0.58 |
| AIFM3 | -0.50 | -0.77 |
| SERPINB6 | -0.50 | -0.70 |
| HLA-F | -0.49 | -0.72 |
| HEPN1 | -0.49 | -0.72 |
| SLC9A3R2 | -0.49 | -0.47 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003676 | nucleic acid binding | IEA | - | |
| GO:0003723 | RNA binding | TAS | 8968755 | |
| GO:0003730 | mRNA 3'-UTR binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 16001084 | |
| GO:0008494 | translation activator activity | IDA | 16001084 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006417 | regulation of translation | IEA | - | |
| GO:0007283 | spermatogenesis | IEA | - | |
| GO:0007281 | germ cell development | TAS | 9288969 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0045948 | positive regulation of translational initiation | IDA | 16001084 | |
| GO:0045836 | positive regulation of meiosis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005844 | polysome | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | TAS | 9288969 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KIM RESPONSE TO TSA AND DECITABINE UP | 129 | 73 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA UP | 183 | 119 | All SZGR 2.0 genes in this pathway |
| MUELLER METHYLATED IN GLIOBLASTOMA | 40 | 22 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL UP | 83 | 51 | All SZGR 2.0 genes in this pathway |
| KLEIN PRIMARY EFFUSION LYMPHOMA UP | 51 | 29 | All SZGR 2.0 genes in this pathway |
| LIANG SILENCED BY METHYLATION 2 | 53 | 34 | All SZGR 2.0 genes in this pathway |
| SU TESTIS | 76 | 53 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED EPIGENETICALLY IN PANCREATIC CANCER | 49 | 30 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS UP | 77 | 57 | All SZGR 2.0 genes in this pathway |
| CHEN ETV5 TARGETS TESTIS | 23 | 14 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| WEBER METHYLATED HCP IN FIBROBLAST DN | 42 | 22 | All SZGR 2.0 genes in this pathway |
| WEBER METHYLATED HCP IN SPERM UP | 20 | 13 | All SZGR 2.0 genes in this pathway |
| MATZUK EMBRYONIC GERM CELL | 19 | 16 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOGONIA | 24 | 17 | All SZGR 2.0 genes in this pathway |
| CONRAD GERMLINE STEM CELL | 13 | 8 | All SZGR 2.0 genes in this pathway |
| SASAI TARGETS OF CXCR6 AND PTCH1 DN | 8 | 6 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 2HR UP | 39 | 30 | All SZGR 2.0 genes in this pathway |
| WAGSCHAL EHMT2 TARGETS UP | 13 | 7 | All SZGR 2.0 genes in this pathway |
| MADAN DPPA4 TARGETS | 46 | 26 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP | 289 | 184 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-140 | 1000 | 1007 | 1A,m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-21 | 719 | 725 | m8 | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-363 | 802 | 808 | 1A | hsa-miR-363 | AUUGCACGGUAUCCAUCUGUAA |
| miR-378* | 561 | 567 | 1A | hsa-miR-422b | CUGGACUUGGAGUCAGAAGGCC |
| hsa-miR-422a | CUGGACUUAGGGUCAGAAGGCC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.