Gene Page: DDC
Summary ?
| GeneID | 1644 |
| Symbol | DDC |
| Synonyms | AADC |
| Description | dopa decarboxylase |
| Reference | MIM:107930|HGNC:HGNC:2719|Ensembl:ENSG00000132437|HPRD:00145|Vega:OTTHUMG00000023353 |
| Gene type | protein-coding |
| Map location | 7p12.2 |
| Pascal p-value | 0.124 |
| Fetal beta | -0.709 |
| eGene | Myers' cis & trans |
| Support | DOPAMINE SEROTONIN |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12066114 | chr1 | 19792526 | DDC | 1644 | 0.17 | trans | ||
| rs17130103 | chr1 | 88150541 | DDC | 1644 | 0.09 | trans | ||
| rs4846645 | chr1 | 220595080 | DDC | 1644 | 0.19 | trans | ||
| rs6666913 | chr1 | 237092144 | DDC | 1644 | 0.01 | trans | ||
| rs13403353 | chr2 | 144067716 | DDC | 1644 | 0.14 | trans | ||
| rs7609917 | chr3 | 76094301 | DDC | 1644 | 0.19 | trans | ||
| rs2324466 | chr3 | 76100753 | DDC | 1644 | 0.01 | trans | ||
| rs17013713 | chr3 | 76105150 | DDC | 1644 | 9.706E-4 | trans | ||
| rs17013722 | chr3 | 76107656 | DDC | 1644 | 0.01 | trans | ||
| rs17013724 | chr3 | 76107684 | DDC | 1644 | 0.01 | trans | ||
| rs13326424 | 0 | DDC | 1644 | 0.16 | trans | |||
| rs11127757 | chr3 | 82043061 | DDC | 1644 | 0.18 | trans | ||
| rs7619146 | chr3 | 82098527 | DDC | 1644 | 0.15 | trans | ||
| rs9986296 | chr5 | 3199318 | DDC | 1644 | 0.01 | trans | ||
| rs200470 | chr6 | 27762186 | DDC | 1644 | 0.12 | trans | ||
| rs839777 | chr6 | 27909656 | DDC | 1644 | 0.13 | trans | ||
| rs149898 | chr6 | 27958071 | DDC | 1644 | 0.13 | trans | ||
| rs156742 | chr6 | 27963191 | DDC | 1644 | 0.13 | trans | ||
| rs175955 | chr6 | 28059733 | DDC | 1644 | 0.13 | trans | ||
| rs1225593 | chr6 | 28150518 | DDC | 1644 | 0.13 | trans | ||
| rs195749 | chr6 | 37253745 | DDC | 1644 | 0.01 | trans | ||
| rs992435 | chr7 | 97050802 | DDC | 1644 | 0.17 | trans | ||
| rs16920368 | chr8 | 55392852 | DDC | 1644 | 0.13 | trans | ||
| rs16927318 | chr8 | 62365911 | DDC | 1644 | 0.03 | trans | ||
| rs180038 | chr9 | 101514415 | DDC | 1644 | 0.11 | trans | ||
| rs4742740 | chr9 | 101524552 | DDC | 1644 | 0.05 | trans | ||
| rs2069410 | chr12 | 56364644 | DDC | 1644 | 8.493E-4 | trans | ||
| rs17834315 | chr13 | 37991091 | DDC | 1644 | 0 | trans | ||
| rs341395 | chr15 | 61127355 | DDC | 1644 | 0.1 | trans | ||
| rs17798004 | chr18 | 70665382 | DDC | 1644 | 0.19 | trans | ||
| rs17736134 | chr18 | 70665805 | DDC | 1644 | 0.18 | trans | ||
| rs2469060 | chr18 | 70673106 | DDC | 1644 | 0.18 | trans | ||
| rs409642 | chr20 | 54250273 | DDC | 1644 | 0.03 | trans | ||
| rs5920627 | chrX | 97385064 | DDC | 1644 | 0.13 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
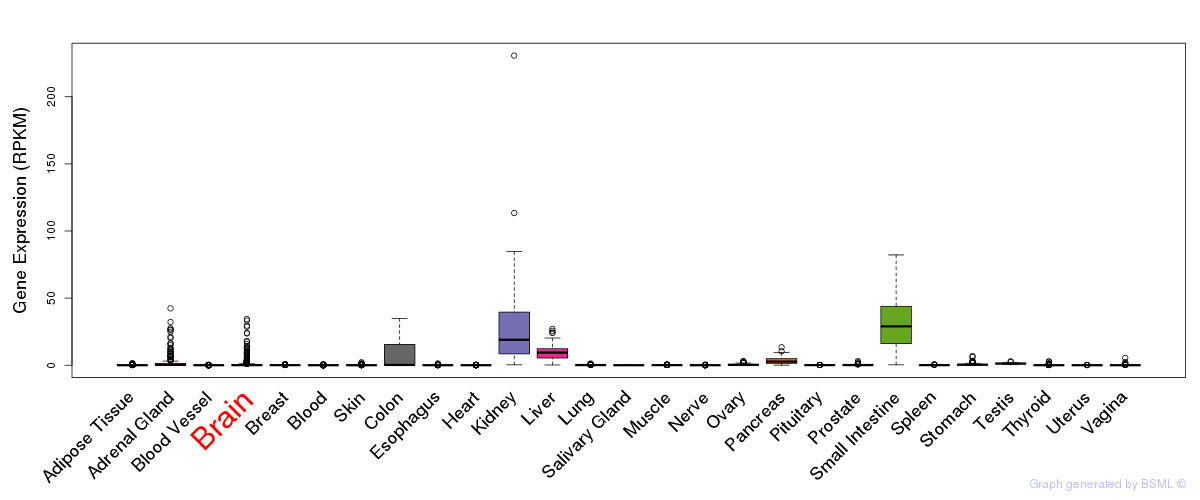
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MED13 | 0.96 | 0.97 |
| MED1 | 0.96 | 0.97 |
| CNOT6 | 0.96 | 0.96 |
| AP1GBP1 | 0.96 | 0.98 |
| AFF4 | 0.96 | 0.96 |
| LATS1 | 0.96 | 0.96 |
| PRKDC | 0.96 | 0.97 |
| ZNF518B | 0.95 | 0.97 |
| PBRM1 | 0.95 | 0.96 |
| NCOA3 | 0.95 | 0.96 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.70 | -0.86 |
| FXYD1 | -0.70 | -0.86 |
| MT-CO2 | -0.70 | -0.87 |
| IFI27 | -0.68 | -0.85 |
| AF347015.27 | -0.68 | -0.83 |
| AIFM3 | -0.68 | -0.74 |
| AF347015.33 | -0.68 | -0.82 |
| S100B | -0.68 | -0.80 |
| HIGD1B | -0.67 | -0.85 |
| HLA-F | -0.67 | -0.73 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004058 | aromatic-L-amino-acid decarboxylase activity | IDA | 16338639 | |
| GO:0005515 | protein binding | IPI | 12864730 | |
| GO:0016829 | lyase activity | IEA | - | |
| GO:0016831 | carboxy-lyase activity | IEA | - | |
| GO:0030170 | pyridoxal phosphate binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006519 | cellular amino acid and derivative metabolic process | IEA | - | |
| GO:0019752 | carboxylic acid metabolic process | IEA | - | |
| GO:0042423 | catecholamine biosynthetic process | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HISTIDINE METABOLISM | 29 | 19 | All SZGR 2.0 genes in this pathway |
| KEGG TYROSINE METABOLISM | 42 | 30 | All SZGR 2.0 genes in this pathway |
| KEGG PHENYLALANINE METABOLISM | 18 | 12 | All SZGR 2.0 genes in this pathway |
| KEGG TRYPTOPHAN METABOLISM | 40 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINE DERIVED HORMONES | 15 | 12 | All SZGR 2.0 genes in this pathway |
| IGARASHI ATF4 TARGETS DN | 90 | 65 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN UP | 184 | 125 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| DIERICK SEROTONIN FUNCTION GENES | 6 | 6 | All SZGR 2.0 genes in this pathway |
| JEON SMAD6 TARGETS DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 2 | 114 | 76 | All SZGR 2.0 genes in this pathway |
| NELSON RESPONSE TO ANDROGEN DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| SMITH LIVER CANCER | 45 | 27 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| ABBUD LIF SIGNALING 1 DN | 26 | 17 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| RORIE TARGETS OF EWSR1 FLI1 FUSION DN | 30 | 23 | All SZGR 2.0 genes in this pathway |
| KUNINGER IGF1 VS PDGFB TARGETS UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| HARRIS BRAIN CANCER PROGENITORS | 44 | 23 | All SZGR 2.0 genes in this pathway |
| LEIN MIDBRAIN MARKERS | 82 | 55 | All SZGR 2.0 genes in this pathway |
| YEGNASUBRAMANIAN PROSTATE CANCER | 128 | 60 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER ERBB2 UP | 147 | 83 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS DN | 109 | 71 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| BRIDEAU IMPRINTED GENES | 63 | 47 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |