Gene Page: AGT
Summary ?
| GeneID | 183 |
| Symbol | AGT |
| Synonyms | ANHU|SERPINA8 |
| Description | angiotensinogen |
| Reference | MIM:106150|HGNC:HGNC:333|Ensembl:ENSG00000135744|HPRD:00106|Vega:OTTHUMG00000037757 |
| Gene type | protein-coding |
| Map location | 1q42.2 |
| Fetal beta | -2.857 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Nucleus accumbens basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1538974 | chr1 | 231828595 | AGT | 183 | 0.17 | cis | ||
| rs11583715 | chr1 | 231829038 | AGT | 183 | 0.17 | cis | ||
| rs16938766 | chr8 | 74949940 | AGT | 183 | 0.19 | trans |
Section II. Transcriptome annotation
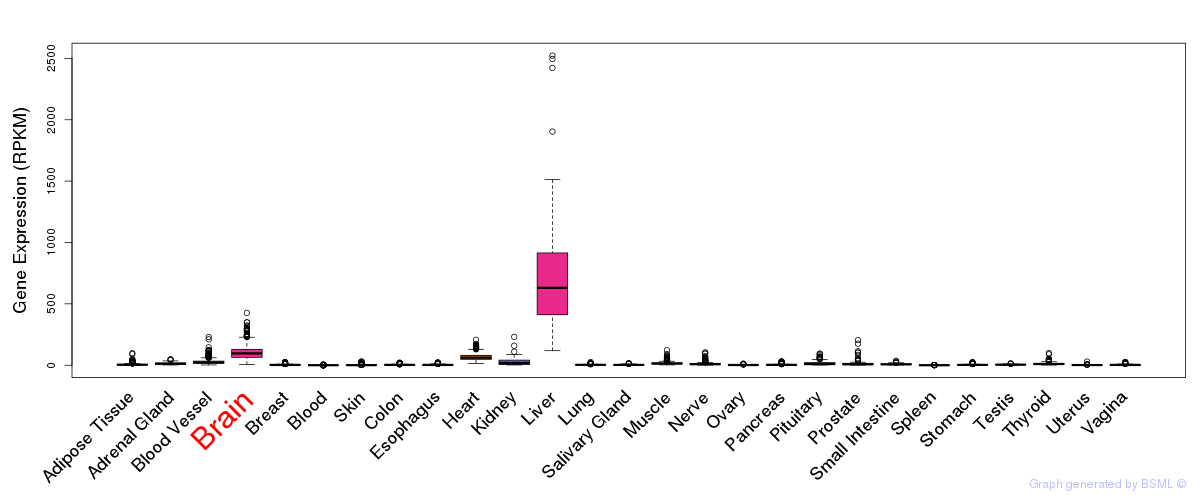
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005179 | hormone activity | IC | 1378723 | |
| GO:0005179 | hormone activity | ISS | - | |
| GO:0004867 | serine-type endopeptidase inhibitor activity | TAS | 3397061 | |
| GO:0008083 | growth factor activity | TAS | 10406457 | |
| GO:0031702 | type 1 angiotensin receptor binding | IEA | - | |
| GO:0031703 | type 2 angiotensin receptor binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008065 | establishment of blood-nerve barrier | IEA | Brain (GO term level: 7) | - |
| GO:0002035 | brain renin-angiotensin system | IEA | Brain (GO term level: 12) | - |
| GO:0048143 | astrocyte activation | IEA | astrocyte (GO term level: 11) | - |
| GO:0043524 | negative regulation of neuron apoptosis | IEA | neuron (GO term level: 9) | - |
| GO:0001568 | blood vessel development | IEA | - | |
| GO:0001558 | regulation of cell growth | NAS | 17159080 | |
| GO:0001543 | ovarian follicle rupture | IEA | - | |
| GO:0001658 | ureteric bud branching | IEA | - | |
| GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure | IEA | - | |
| GO:0001999 | renal response to blood flow during renin-angiotensin regulation of systemic arterial blood pressure | IEA | - | |
| GO:0002018 | renin-angiotensin regulation of aldosterone production | IEA | - | |
| GO:0002019 | regulation of renal output by angiotensin | IEA | - | |
| GO:0003078 | regulation of natriuresis | NAS | 17159080 | |
| GO:0001822 | kidney development | IEA | - | |
| GO:0001974 | blood vessel remodeling | TAS | 10406457 | |
| GO:0001819 | positive regulation of cytokine production | TAS | 17906677 | |
| GO:0007200 | G-protein signaling, coupled to IP3 second messenger (phospholipase C activating) | NAS | 17159080 | |
| GO:0007160 | cell-matrix adhesion | IEA | - | |
| GO:0007199 | G-protein signaling, coupled to cGMP nucleotide second messenger | TAS | 17159080 | |
| GO:0007267 | cell-cell signaling | TAS | 8513325 | |
| GO:0007263 | nitric oxide mediated signal transduction | TAS | 17159080 | |
| GO:0010595 | positive regulation of endothelial cell migration | IDA | 15652490 | |
| GO:0008285 | negative regulation of cell proliferation | IEA | - | |
| GO:0009409 | response to cold | IEA | - | |
| GO:0010613 | positive regulation of cardiac muscle hypertrophy | ISS | - | |
| GO:0006800 | oxygen and reactive oxygen species metabolic process | TAS | 17159080 |17906677 | |
| GO:0014068 | positive regulation of phosphoinositide 3-kinase cascade | IDA | 15652490 | |
| GO:0043065 | positive regulation of apoptosis | IDA | 10406457 | |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | TAS | 17906677 | |
| GO:0014873 | response to muscle activity involved in regulation of muscle adaptation | ISS | - | |
| GO:0030432 | peristalsis | IEA | - | |
| GO:0042445 | hormone metabolic process | IEA | - | |
| GO:0042756 | drinking behavior | IEA | - | |
| GO:0040018 | positive regulation of multicellular organism growth | IEA | - | |
| GO:0019229 | regulation of vasoconstriction | NAS | 17159080 | |
| GO:0033864 | positive regulation of NAD(P)H oxidase activity | TAS | 17159080 | |
| GO:0051145 | smooth muscle cell differentiation | IEA | - | |
| GO:0043410 | positive regulation of MAPKKK cascade | IEA | - | |
| GO:0030198 | extracellular matrix organization | IEA | - | |
| GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway | IDA | 15652490 | |
| GO:0045723 | positive regulation of fatty acid biosynthetic process | IEA | - | |
| GO:0048146 | positive regulation of fibroblast proliferation | ISS | - | |
| GO:0050729 | positive regulation of inflammatory response | TAS | 17159080 |17906677 | |
| GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | IDA | 15652490 | |
| GO:0046622 | positive regulation of organ growth | IEA | - | |
| GO:0048659 | smooth muscle cell proliferation | IEA | - | |
| GO:0051387 | negative regulation of nerve growth factor receptor signaling pathway | IDA | 10406457 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | NAS | 14718574 | |
| GO:0005615 | extracellular space | IEA | - | |
| GO:0005625 | soluble fraction | TAS | 6089875 | |
| GO:0005737 | cytoplasm | IDA | 18029348 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RENIN ANGIOTENSIN SYSTEM | 17 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AT1R PATHWAY | 36 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ACE2 PATHWAY | 13 | 7 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BIOPEPTIDES PATHWAY | 45 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFAT PATHWAY | 56 | 45 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARDIACEGF PATHWAY | 18 | 16 | All SZGR 2.0 genes in this pathway |
| PID PRL SIGNALING EVENTS PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Q SIGNALLING EVENTS | 184 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ MITOTIC ARREST BY DOCETAXEL 1 UP | 36 | 18 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 2NM UP | 81 | 57 | All SZGR 2.0 genes in this pathway |
| DAUER STAT3 TARGETS UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 DN | 51 | 42 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| ZEILSTRA CD44 TARGETS UP | 7 | 5 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS | 183 | 115 | All SZGR 2.0 genes in this pathway |
| STOSSI RESPONSE TO ESTRADIOL | 50 | 35 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY DN | 48 | 34 | All SZGR 2.0 genes in this pathway |
| RADMACHER AML PROGNOSIS | 78 | 52 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| MARCHINI TRABECTEDIN RESISTANCE UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF DN | 84 | 50 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 5 | 126 | 78 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 24HR | 43 | 30 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| LI ADIPOGENESIS BY ACTIVATED PPARG | 17 | 12 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS UP | 49 | 40 | All SZGR 2.0 genes in this pathway |
| LEIN MIDBRAIN MARKERS | 82 | 55 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SCHRAETS MLL TARGETS DN | 33 | 24 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 DN | 80 | 51 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC LCP WITH H3K4ME3 | 58 | 34 | All SZGR 2.0 genes in this pathway |
| GERHOLD RESPONSE TO TZD DN | 13 | 11 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| VERNOCHET ADIPOGENESIS | 19 | 11 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |