Gene Page: EFNB1
Summary ?
| GeneID | 1947 |
| Symbol | EFNB1 |
| Synonyms | CFND|CFNS|EFB1|EFL3|EPLG2|Elk-L|LERK2 |
| Description | ephrin-B1 |
| Reference | MIM:300035|HGNC:HGNC:3226|Ensembl:ENSG00000090776|HPRD:02072|Vega:OTTHUMG00000021751 |
| Gene type | protein-coding |
| Map location | Xq12 |
| Fetal beta | 0.993 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:PGC128 | Genome-wide Association Study | A multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls. Reported by the Schizophrenia Working Group of PGC. 128 independent associations spanning 108 loci | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0172 |
Section I. Genetics and epigenetics annotation
 CV:PGC128
CV:PGC128
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs5937157 | chrX | 68377126 | TG | 5.742E-11 | intergenic | EFNB1,PJA1 | dist=315120;dist=3455 |
Section II. Transcriptome annotation
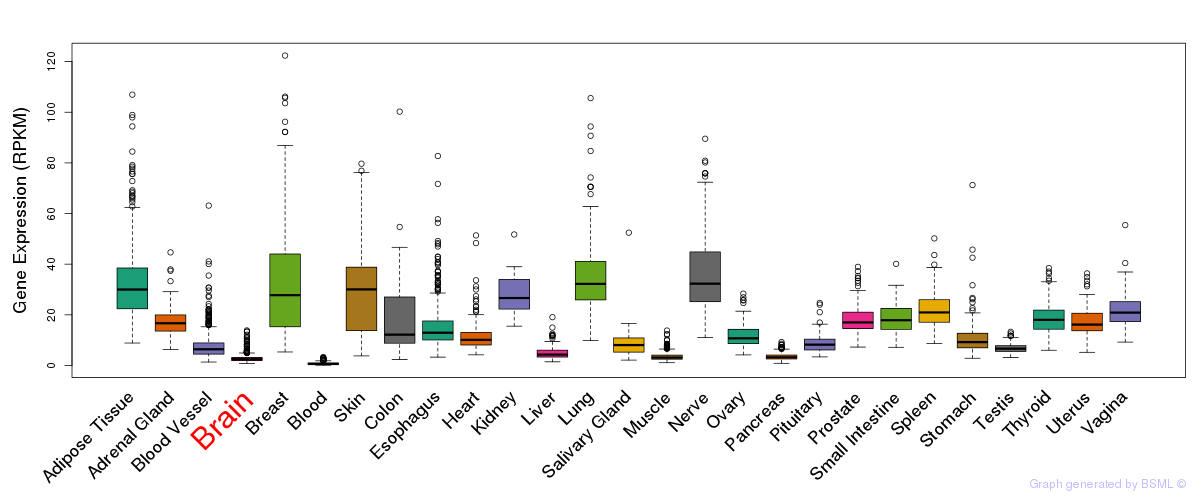
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GRIN2A | 0.91 | 0.86 |
| DLGAP2 | 0.90 | 0.82 |
| MEF2D | 0.89 | 0.74 |
| DIRAS2 | 0.89 | 0.77 |
| TMEM132D | 0.88 | 0.82 |
| KCNB1 | 0.88 | 0.76 |
| IQSEC2 | 0.88 | 0.82 |
| TRHDE | 0.88 | 0.86 |
| MFSD4 | 0.87 | 0.88 |
| PITPNM3 | 0.87 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.46 | -0.57 |
| HEBP2 | -0.46 | -0.66 |
| GTF3C6 | -0.43 | -0.45 |
| RPL23A | -0.43 | -0.52 |
| FADS2 | -0.43 | -0.39 |
| C9orf46 | -0.42 | -0.48 |
| C21orf57 | -0.42 | -0.49 |
| TRAF4 | -0.41 | -0.50 |
| RPS12 | -0.41 | -0.56 |
| DYNLT1 | -0.40 | -0.56 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 9883737 | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0046875 | ephrin receptor binding | TAS | 8798744 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007411 | axon guidance | IEA | axon (GO term level: 13) | - |
| GO:0007399 | nervous system development | IEA | neurite (GO term level: 5) | - |
| GO:0001755 | neural crest cell migration | IEA | - | |
| GO:0007155 | cell adhesion | TAS | 8798744 | |
| GO:0007267 | cell-cell signaling | TAS | 8798744 | |
| GO:0009880 | embryonic pattern specification | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0042102 | positive regulation of T cell proliferation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045202 | synapse | ISS | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005625 | soluble fraction | TAS | 8798744 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 8660976 | |
| GO:0045121 | membrane raft | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| EPHA3 | ETK | ETK1 | HEK | HEK4 | TYRO4 | EPH receptor A3 | Reconstituted Complex | BioGRID | 9195962 |
| EPHB1 | ELK | EPHT2 | FLJ37986 | Hek6 | NET | EPH receptor B1 | The membrane bound ligand ELK-L interacts with the receptor ELK. This interaction was modeled on a demonstrated interaction between ELK-L from human and ELK from rat. | BIND | 7973638 |8755474 |
| EPHB1 | ELK | EPHT2 | FLJ37986 | Hek6 | NET | EPH receptor B1 | - | HPRD,BioGRID | 10669731 |12223469 |
| EPHB2 | CAPB | DRT | EPHT3 | ERK | Hek5 | MGC87492 | PCBC | Tyro5 | EPH receptor B2 | - | HPRD | 8878483 |
| EPHB2 | CAPB | DRT | EPHT3 | ERK | Hek5 | MGC87492 | PCBC | Tyro5 | EPH receptor B2 | The membrane bound ligand ELK-L interacts with the receptor NUK. This interaction was modeled on a demonstrated interaction between ELK-L from human and NUK from mouse. | BIND | 8755474 |
| GRIP2 | - | glutamate receptor interacting protein 2 | in vitro in vivo Two-hybrid | BioGRID | 9883737 |10197531 |
| NCK2 | GRB4 | NCKbeta | NCK adaptor protein 2 | - | HPRD | 11557983 |
| PICK1 | MGC15204 | PICK | PRKCABP | protein interacting with PRKCA 1 | - | HPRD,BioGRID | 9883737 |
| PTPN13 | DKFZp686J1497 | FAP-1 | PNP1 | PTP-BAS | PTP-BL | PTP1E | PTPL1 | PTPLE | protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) | - | HPRD,BioGRID | 9920925 |
| RGS3 | C2PA | FLJ20370 | FLJ31516 | FLJ90496 | PDZ-RGS3 | RGP3 | regulator of G-protein signaling 3 | - | HPRD,BioGRID | 11301003 |
| SDCBP | MDA-9 | ST1 | SYCL | TACIP18 | syndecan binding protein (syntenin) | - | HPRD,BioGRID | 9920925 |
| SORBS1 | CAP | DKFZp451C066 | DKFZp586P1422 | FLAF2 | FLJ12406 | KIAA1296 | R85FL | SH3D5 | SH3P12 | SORB1 | sorbin and SH3 domain containing 1 | - | HPRD | 11557983 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 8878483 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID EPHB FWD PATHWAY | 40 | 38 | All SZGR 2.0 genes in this pathway |
| PID EPHRINB REV PATHWAY | 30 | 25 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION DN | 100 | 64 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL UP | 83 | 51 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL FGF3 | 8 | 7 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| GENTILE UV LOW DOSE UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS DN | 53 | 34 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS DN | 135 | 88 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS DN | 25 | 21 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY GAMMA IN WS | 33 | 22 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE UP | 56 | 41 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| HOFFMANN SMALL PRE BII TO IMMATURE B LYMPHOCYTE DN | 50 | 33 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| TERAO AOX4 TARGETS SKIN UP | 38 | 27 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 834 | 840 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-124/506 | 200 | 206 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-129-5p | 730 | 736 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-140 | 1496 | 1502 | 1A | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-15/16/195/424/497 | 1178 | 1184 | 1A | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-150 | 1303 | 1309 | 1A | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-17-5p/20/93.mr/106/519.d | 1148 | 1154 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-185 | 936 | 942 | m8 | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-208 | 799 | 805 | 1A | hsa-miR-208 | AUAAGACGAGCAAAAAGCUUGU |
| miR-34/449 | 1189 | 1195 | 1A | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-381 | 1425 | 1431 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-491 | 63 | 69 | 1A | hsa-miR-491brain | AGUGGGGAACCCUUCCAUGAGGA |
| miR-496 | 1412 | 1418 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-499 | 799 | 805 | 1A | hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA |
| miR-503 | 1178 | 1184 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.