Gene Page: EGR3
Summary ?
| GeneID | 1960 |
| Symbol | EGR3 |
| Synonyms | EGR-3|PILOT |
| Description | early growth response 3 |
| Reference | MIM:602419|HGNC:HGNC:3240|Ensembl:ENSG00000179388|HPRD:03881|Vega:OTTHUMG00000097825 |
| Gene type | protein-coding |
| Map location | 8p23-p21 |
| Pascal p-value | 0.773 |
| Fetal beta | -4.408 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.03086 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.00057 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs5944689 | chrX | 24860329 | EGR3 | 1960 | 0.14 | trans | ||
| rs5973369 | chrX | 35392270 | EGR3 | 1960 | 0.17 | trans | ||
| rs5973375 | chrX | 35489060 | EGR3 | 1960 | 0.2 | trans | ||
| rs2171999 | chrX | 35498985 | EGR3 | 1960 | 0.19 | trans |
Section II. Transcriptome annotation
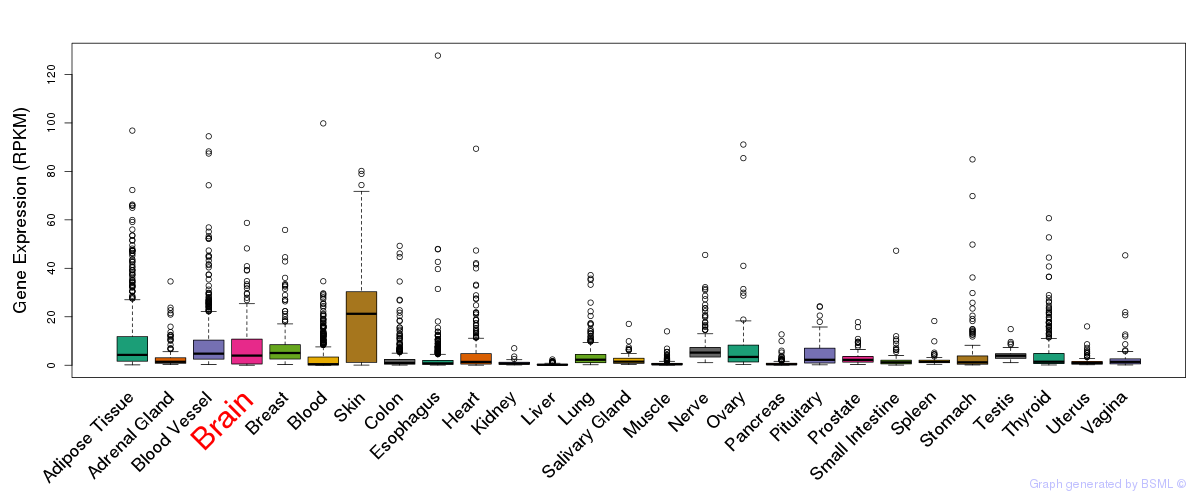
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NONO | 0.96 | 0.93 |
| INTS7 | 0.96 | 0.95 |
| XRN2 | 0.96 | 0.93 |
| EIF2S3 | 0.96 | 0.95 |
| RBMX | 0.96 | 0.93 |
| CSGALNACT2 | 0.95 | 0.94 |
| USP3 | 0.95 | 0.82 |
| MCPH1 | 0.95 | 0.94 |
| ANP32A | 0.95 | 0.93 |
| CAPRIN1 | 0.95 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AIFM3 | -0.69 | -0.83 |
| HLA-F | -0.69 | -0.79 |
| C5orf53 | -0.68 | -0.76 |
| PTH1R | -0.67 | -0.78 |
| FXYD1 | -0.67 | -0.91 |
| AF347015.31 | -0.67 | -0.87 |
| IFI27 | -0.67 | -0.90 |
| TSC22D4 | -0.67 | -0.82 |
| MT-CO2 | -0.66 | -0.88 |
| HEPN1 | -0.66 | -0.77 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 1906159 |9731539 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007274 | neuromuscular synaptic transmission | IEA | neuron, Synap (GO term level: 7) | - |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007517 | muscle development | TAS | 9731539 | |
| GO:0007623 | circadian rhythm | TAS | 9488654 | |
| GO:0007422 | peripheral nervous system development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA VIP PATHWAY | 29 | 26 | All SZGR 2.0 genes in this pathway |
| ST DIFFERENTIATION PATHWAY IN PC12 CELLS | 45 | 35 | All SZGR 2.0 genes in this pathway |
| PID NFAT TFPATHWAY | 47 | 39 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL DN | 60 | 39 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA EGF SIGNALING UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| PACHER TARGETS OF IGF1 AND IGF2 UP | 35 | 27 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS E UP | 97 | 60 | All SZGR 2.0 genes in this pathway |
| CHASSOT SKIN WOUND | 10 | 9 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY | 66 | 48 | All SZGR 2.0 genes in this pathway |
| THEODOROU MAMMARY TUMORIGENESIS | 31 | 24 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 40 HELA | 42 | 29 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 40 MCF10A | 19 | 11 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 60 MCF10A | 57 | 42 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT | 108 | 73 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL UP | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BECKER TAMOXIFEN RESISTANCE UP | 50 | 36 | All SZGR 2.0 genes in this pathway |
| KUROKAWA LIVER CANCER CHEMOTHERAPY DN | 41 | 28 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS UP | 48 | 34 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND UP | 77 | 52 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT DN | 102 | 67 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| ABE VEGFA TARGETS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| ABE VEGFA TARGETS 30MIN | 29 | 21 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| MASSARWEH RESPONSE TO ESTRADIOL | 61 | 47 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS DN | 68 | 48 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS UP | 77 | 56 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| LEE SP4 THYMOCYTE | 14 | 11 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| ZHAN EARLY DIFFERENTIATION GENES DN | 42 | 29 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| UZONYI RESPONSE TO LEUKOTRIENE AND THROMBIN | 37 | 26 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 12HR UP | 29 | 18 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 24HR DN | 43 | 32 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 32HR DN | 39 | 28 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 48HR DN | 40 | 29 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN UP | 56 | 38 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY TARGETS OF MUTATED TP53 UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP | 163 | 111 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 1 | 46 | 33 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 1HR UP | 34 | 26 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR DN | 114 | 69 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 2 TRANSIENTLY INDUCED BY EGF | 51 | 29 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 1594 | 1601 | 1A,m8 | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-142-5p | 1827 | 1833 | m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-146 | 2250 | 2256 | 1A | hsa-miR-146a | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-146bbrain | UGAGAACUGAAUUCCAUAGGCU | ||||
| miR-17-5p/20/93.mr/106/519.d | 1826 | 1832 | 1A | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-181 | 1724 | 1731 | 1A,m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-182 | 2291 | 2298 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-200bc/429 | 2077 | 2083 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-203.1 | 2590 | 2597 | 1A,m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-216 | 1359 | 1366 | 1A,m8 | hsa-miR-216 | UAAUCUCAGCUGGCAACUGUG |
| miR-219 | 2741 | 2748 | 1A,m8 | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-23 | 2376 | 2382 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC | ||||
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-30-5p | 1800 | 1806 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-323 | 2101 | 2108 | 1A,m8 | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-331 | 289 | 295 | m8 | hsa-miR-331brain | GCCCCUGGGCCUAUCCUAGAA |
| miR-335 | 581 | 587 | 1A | hsa-miR-335brain | UCAAGAGCAAUAACGAAAAAUGU |
| miR-380-5p | 1014 | 1020 | 1A | hsa-miR-380-5p | UGGUUGACCAUAGAACAUGCGC |
| hsa-miR-563 | AGGUUGACAUACGUUUCCC | ||||
| miR-381 | 2448 | 2454 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-494 | 712 | 718 | m8 | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
| miR-7 | 690 | 697 | 1A,m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
| miR-9 | 438 | 444 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-96 | 2292 | 2298 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.