Gene Page: PAOX
Summary ?
| GeneID | 196743 |
| Symbol | PAOX |
| Synonyms | PAO |
| Description | polyamine oxidase (exo-N4-amino) |
| Reference | MIM:615853|HGNC:HGNC:20837|Ensembl:ENSG00000148832|HPRD:10136|Vega:OTTHUMG00000019318 |
| Gene type | protein-coding |
| Map location | 10q26.3 |
| Pascal p-value | 0.045 |
| Sherlock p-value | 0.692 |
| Fetal beta | -0.338 |
| DMG | 1 (# studies) |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.03487 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17465130 | 10 | 135192054 | PAOX | 1.17E-10 | -0.017 | 4.75E-7 | DMG:Jaffe_2016 |
| cg19601447 | 10 | 135192230 | PAOX | 3.25E-8 | -0.012 | 9.77E-6 | DMG:Jaffe_2016 |
| cg12292686 | 10 | 135192102 | PAOX | 6.53E-8 | -0.011 | 1.61E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
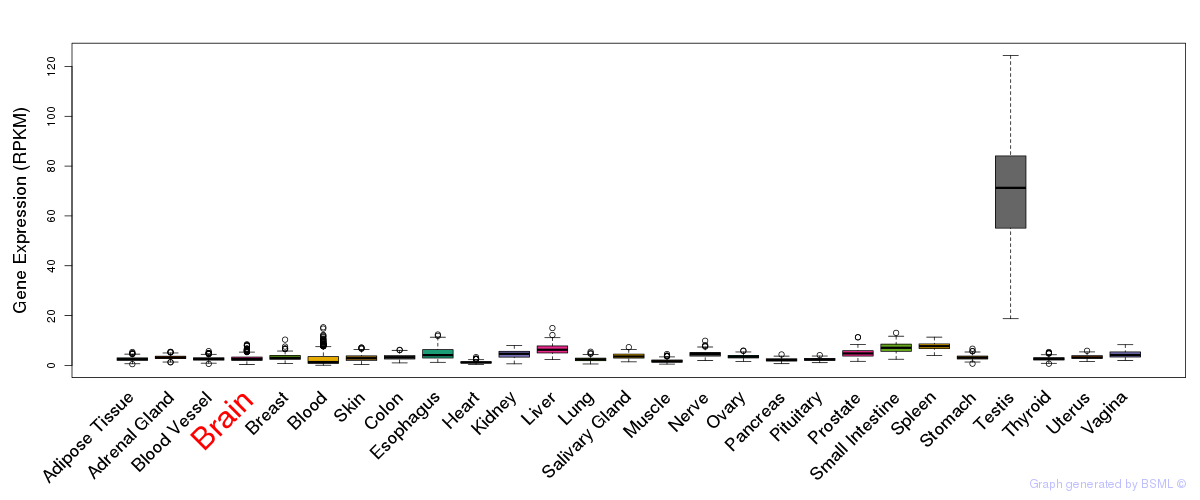
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PEROXISOME | 78 | 47 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A9B1 PATHWAY | 25 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME BIOLOGICAL OXIDATIONS | 139 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | 70 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF POLYAMINES | 15 | 11 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS UP | 88 | 47 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER POOR SURVIVAL DN | 22 | 15 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE DN | 80 | 54 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |