Gene Page: EIF4A2
Summary ?
| GeneID | 1974 |
| Symbol | EIF4A2 |
| Synonyms | BM-010|DDX2B|EIF4A|EIF4F|eIF-4A-II|eIF4A-II |
| Description | eukaryotic translation initiation factor 4A2 |
| Reference | MIM:601102|HGNC:HGNC:3284|Ensembl:ENSG00000156976|HPRD:03062|Vega:OTTHUMG00000156564 |
| Gene type | protein-coding |
| Map location | 3q28 |
| Pascal p-value | 0.099 |
| Sherlock p-value | 0.002 |
| Fetal beta | -0.338 |
| eGene | Myers' cis & trans |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7132043 | chr12 | 80968399 | EIF4A2 | 1974 | 0.09 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
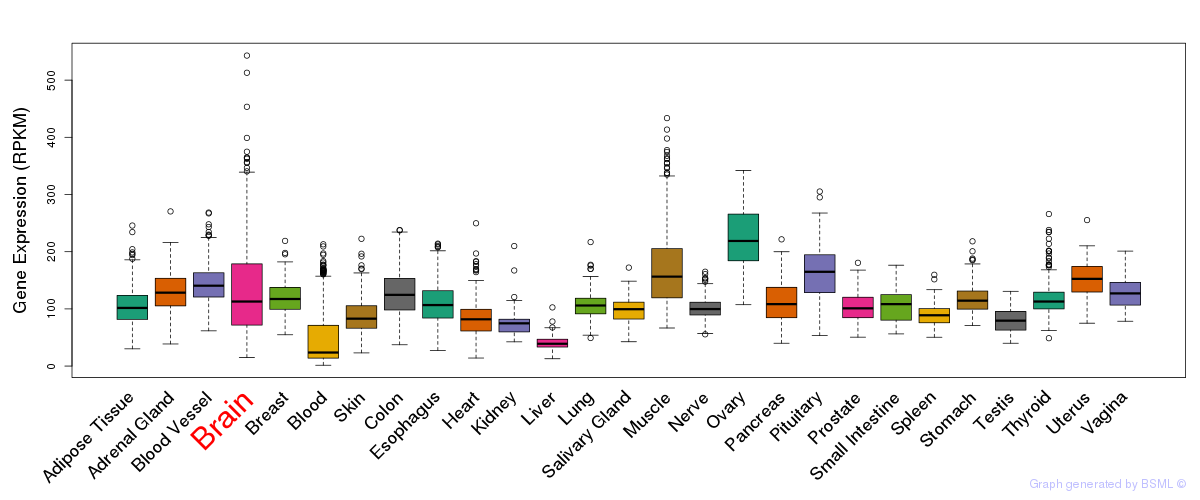
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003676 | nucleic acid binding | IEA | - | |
| GO:0003723 | RNA binding | IEA | - | |
| GO:0003743 | translation initiation factor activity | IEA | - | |
| GO:0003743 | translation initiation factor activity | TAS | 8521730 | |
| GO:0005515 | protein binding | IPI | 11408474 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0008026 | ATP-dependent helicase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006446 | regulation of translational initiation | TAS | 8521730 | |
| GO:0006412 | translation | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 8449919 |12588972 | |
| GO:0016281 | eukaryotic translation initiation factor 4F complex | TAS | 8521730 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ALPI | IAP | alkaline phosphatase, intestinal | Affinity Capture-MS | BioGRID | 17353931 |
| ATRX | ATR2 | MGC2094 | MRXHF1 | RAD54 | RAD54L | SFM1 | SHS | XH2 | XNP | ZNF-HX | alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| CD2BP2 | FWP010 | LIN1 | Snu40 | U5-52K | CD2 (cytoplasmic tail) binding protein 2 | Affinity Capture-MS | BioGRID | 17353931 |
| CMBL | FLJ23617 | carboxymethylenebutenolidase homolog (Pseudomonas) | Affinity Capture-MS | BioGRID | 17353931 |
| DDX3X | DBX | DDX14 | DDX3 | HLP2 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3A | EIF3 | EIF3S10 | KIAA0139 | P167 | TIF32 | eIF3-p170 | eIF3-theta | p180 | p185 | eukaryotic translation initiation factor 3, subunit A | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3D | EIF3S7 | MGC126526 | MGC17258 | eIF3-p66 | eIF3-zeta | eukaryotic translation initiation factor 3, subunit D | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3EIP | EIF3S11 | EIF3S6IP | HSPC021 | HSPC025 | MSTP005 | eukaryotic translation initiation factor 3, subunit E interacting protein | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3H | EIF3S3 | MGC102958 | eIF3-gamma | eIF3-p40 | eukaryotic translation initiation factor 3, subunit H | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3K | ARG134 | EIF3-p28 | EIF3S12 | HSPC029 | M9 | MSTP001 | PLAC-24 | PLAC24 | PRO1474 | PTD001 | eukaryotic translation initiation factor 3, subunit K | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3M | B5 | FLJ29030 | GA17 | PCID1 | hfl-B5 | eukaryotic translation initiation factor 3, subunit M | Affinity Capture-MS | BioGRID | 17353931 |
| EIF4E | CBP | EIF4E1 | EIF4EL1 | EIF4F | MGC111573 | eukaryotic translation initiation factor 4E | Affinity Capture-MS | BioGRID | 17353931 |
| EIF4G1 | DKFZp686A1451 | EIF4F | EIF4G | p220 | eukaryotic translation initiation factor 4 gamma, 1 | Affinity Capture-MS | BioGRID | 17353931 |
| EIF4G1 | DKFZp686A1451 | EIF4F | EIF4G | p220 | eukaryotic translation initiation factor 4 gamma, 1 | - | HPRD | 11408474 |
| EIF4G2 | AAG1 | DAP5 | FLJ41344 | NAT1 | p97 | eukaryotic translation initiation factor 4 gamma, 2 | Affinity Capture-MS | BioGRID | 17353931 |
| EIF4G3 | eIF4GII | eukaryotic translation initiation factor 4 gamma, 3 | Affinity Capture-MS | BioGRID | 17353931 |
| MYO1B | myr1 | myosin IB | Affinity Capture-MS | BioGRID | 17353931 |
| P4HB | DSI | ERBA2L | GIT | PDI | PDIA1 | PHDB | PO4DB | PO4HB | PROHB | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| PABPC1 | PAB1 | PABP | PABP1 | PABPC2 | PABPL1 | poly(A) binding protein, cytoplasmic 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PDCD4 | H731 | MGC33046 | MGC33047 | programmed cell death 4 (neoplastic transformation inhibitor) | Affinity Capture-MS | BioGRID | 17353931 |
| PTPRS | PTPSIGMA | protein tyrosine phosphatase, receptor type, S | Affinity Capture-MS | BioGRID | 17353931 |
| RPS29 | - | ribosomal protein S29 | Affinity Capture-MS | BioGRID | 17353931 |
| SFRS17A | 721P | CCDC133 | CXYorf3 | DXYS155E | MGC125365 | MGC125366 | MGC39904 | XE7 | XE7Y | splicing factor, arginine/serine-rich 17A | Affinity Capture-MS | BioGRID | 17353931 |
| SPAG9 | FLJ13450 | FLJ14006 | FLJ26141 | FLJ34602 | HLC4 | JLP | KIAA0516 | MGC117291 | MGC14967 | MGC74461 | PHET | PIG6 | sperm associated antigen 9 | Affinity Capture-MS | BioGRID | 17353931 |
| TPM3 | FLJ41118 | MGC14582 | MGC3261 | MGC72094 | NEM1 | OK/SW-cl.5 | TM-5 | TM3 | TM30 | TM30nm | TPMsk3 | TRK | hscp30 | tropomyosin 3 | Affinity Capture-MS | BioGRID | 17353931 |
| VAC14 | ArPIKfyve | FLJ10305 | FLJ36622 | FLJ46582 | MGC149815 | MGC149816 | TAX1BP2 | TRX | Vac14 homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA EIF PATHWAY | 16 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MTOR PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EIF4 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSLATION | 222 | 75 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | 66 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | 84 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | 176 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME DEADENYLATION OF MRNA | 22 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME DEADENYLATION DEPENDENT MRNA DECAY | 48 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION CCNE1 | 40 | 26 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 7 | 8 | 6 | All SZGR 2.0 genes in this pathway |
| ZHAN V2 LATE DIFFERENTIATION GENES | 45 | 34 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| MMS MOUSE LYMPH HIGH 4HRS UP | 36 | 20 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| LEE AGING MUSCLE UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D UP | 69 | 46 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE E2 | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| CHNG MULTIPLE MYELOMA HYPERPLOID UP | 52 | 25 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S2 | 115 | 74 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-132/212 | 202 | 208 | m8 | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-137 | 531 | 537 | m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-145 | 504 | 510 | m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-17-5p/20/93.mr/106/519.d | 620 | 626 | 1A | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-181 | 596 | 602 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-183 | 567 | 573 | 1A | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-19 | 574 | 580 | 1A | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-208 | 582 | 588 | 1A | hsa-miR-208 | AUAAGACGAGCAAAAAGCUUGU |
| miR-217 | 22 | 28 | m8 | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-335 | 159 | 165 | m8 | hsa-miR-335brain | UCAAGAGCAAUAACGAAAAAUGU |
| miR-377 | 499 | 505 | 1A | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-381 | 611 | 617 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-499 | 582 | 588 | 1A | hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA |
| miR-543 | 597 | 604 | 1A,m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.