Gene Page: ALAS2
Summary ?
| GeneID | 212 |
| Symbol | ALAS2 |
| Synonyms | ALAS-E|ALASE|ANH1|ASB|XLDPP|XLEPP|XLSA |
| Description | 5'-aminolevulinate synthase 2 |
| Reference | MIM:301300|HGNC:HGNC:397|Ensembl:ENSG00000158578|HPRD:02356|Vega:OTTHUMG00000021641 |
| Gene type | protein-coding |
| Map location | Xp11.21 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2762819 | chr1 | 52798032 | ALAS2 | 212 | 0.15 | trans | ||
| rs7538562 | chr1 | 59277182 | ALAS2 | 212 | 0.04 | trans | ||
| rs2811849 | chr1 | 59300272 | ALAS2 | 212 | 0.14 | trans | ||
| rs2811850 | chr1 | 59312340 | ALAS2 | 212 | 0.14 | trans | ||
| rs2272792 | chr1 | 166829521 | ALAS2 | 212 | 1.638E-4 | trans | ||
| rs6543264 | chr2 | 105270432 | ALAS2 | 212 | 2.924E-5 | trans | ||
| rs12468303 | chr2 | 181613256 | ALAS2 | 212 | 0.11 | trans | ||
| rs16867364 | chr2 | 181624945 | ALAS2 | 212 | 0.2 | trans | ||
| rs6442062 | chr3 | 47243234 | ALAS2 | 212 | 0.12 | trans | ||
| rs295432 | chr3 | 47361887 | ALAS2 | 212 | 0 | trans | ||
| rs1545329 | chr4 | 95912889 | ALAS2 | 212 | 8.034E-4 | trans | ||
| rs2120832 | chr4 | 95924906 | ALAS2 | 212 | 0.02 | trans | ||
| rs1444920 | chr4 | 95925993 | ALAS2 | 212 | 0.02 | trans | ||
| rs9326981 | chr5 | 115320607 | ALAS2 | 212 | 0.1 | trans | ||
| rs12524113 | chr6 | 38679547 | ALAS2 | 212 | 5.033E-7 | trans | ||
| rs17712118 | chr6 | 70808553 | ALAS2 | 212 | 0.1 | trans | ||
| rs9484130 | chr6 | 138547963 | ALAS2 | 212 | 0.04 | trans | ||
| rs6907613 | chr6 | 138548270 | ALAS2 | 212 | 0.09 | trans | ||
| rs11007270 | chr10 | 29146063 | ALAS2 | 212 | 0.16 | trans | ||
| rs266080 | chr10 | 44833893 | ALAS2 | 212 | 0.17 | trans | ||
| rs7488206 | chr12 | 49784155 | ALAS2 | 212 | 0.13 | trans | ||
| rs12422793 | chr12 | 125892834 | ALAS2 | 212 | 0.09 | trans | ||
| rs7323925 | chr13 | 24217159 | ALAS2 | 212 | 0.19 | trans | ||
| snp_a-1785245 | 0 | ALAS2 | 212 | 0.05 | trans | |||
| rs4566169 | chr16 | 911716 | ALAS2 | 212 | 0.15 | trans | ||
| rs274094 | chr16 | 24910476 | ALAS2 | 212 | 0.15 | trans | ||
| rs6029091 | chr20 | 39016259 | ALAS2 | 212 | 0.11 | trans |
Section II. Transcriptome annotation
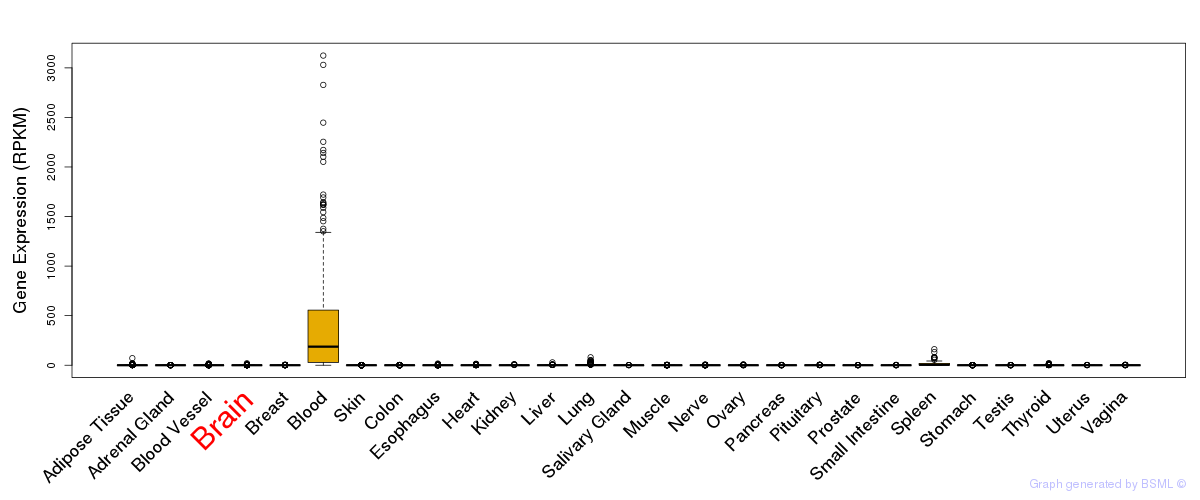
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TELO2 | 0.90 | 0.89 |
| ARVCF | 0.90 | 0.88 |
| DTX1 | 0.90 | 0.89 |
| WIZ | 0.89 | 0.89 |
| FZR1 | 0.89 | 0.87 |
| GLT25D1 | 0.89 | 0.87 |
| ZNF446 | 0.89 | 0.90 |
| SARM1 | 0.88 | 0.86 |
| RFX1 | 0.88 | 0.90 |
| GTPBP3 | 0.88 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.70 | -0.70 |
| AF347015.31 | -0.68 | -0.74 |
| AF347015.27 | -0.67 | -0.73 |
| MT-CO2 | -0.65 | -0.72 |
| AF347015.33 | -0.65 | -0.71 |
| MT-CYB | -0.63 | -0.70 |
| S100B | -0.63 | -0.67 |
| ACOT13 | -0.62 | -0.65 |
| AF347015.21 | -0.62 | -0.77 |
| B2M | -0.61 | -0.65 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCINE SERINE AND THREONINE METABOLISM | 31 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG PORPHYRIN AND CHLOROPHYLL METABOLISM | 41 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AHSP PATHWAY | 13 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PORPHYRINS | 14 | 7 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY UP | 78 | 41 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP | 98 | 58 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK DN | 79 | 54 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA DN | 65 | 44 | All SZGR 2.0 genes in this pathway |
| WELCH GATA1 TARGETS | 22 | 12 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGG VS IGA DN | 26 | 16 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS MATURE CELL | 293 | 160 | All SZGR 2.0 genes in this pathway |
| KEEN RESPONSE TO ROSIGLITAZONE DN | 106 | 68 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 DN | 40 | 22 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN | 242 | 146 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS DN | 116 | 74 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS NORMAL EMBRYOS DN | 25 | 13 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 UP | 115 | 73 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |