Gene Page: EXTL1
Summary ?
| GeneID | 2134 |
| Symbol | EXTL1 |
| Synonyms | EXTL |
| Description | exostosin-like glycosyltransferase 1 |
| Reference | MIM:601738|HGNC:HGNC:3515|Ensembl:ENSG00000158008|HPRD:03441|Vega:OTTHUMG00000007509 |
| Gene type | protein-coding |
| Map location | 1p36.1 |
| Pascal p-value | 0.514 |
| Sherlock p-value | 0.974 |
| Fetal beta | -1.076 |
| eGene | Cerebellum |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EXTL1 | chr1 | 26360265 | G | A | NM_004455 | p.533E>K | missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
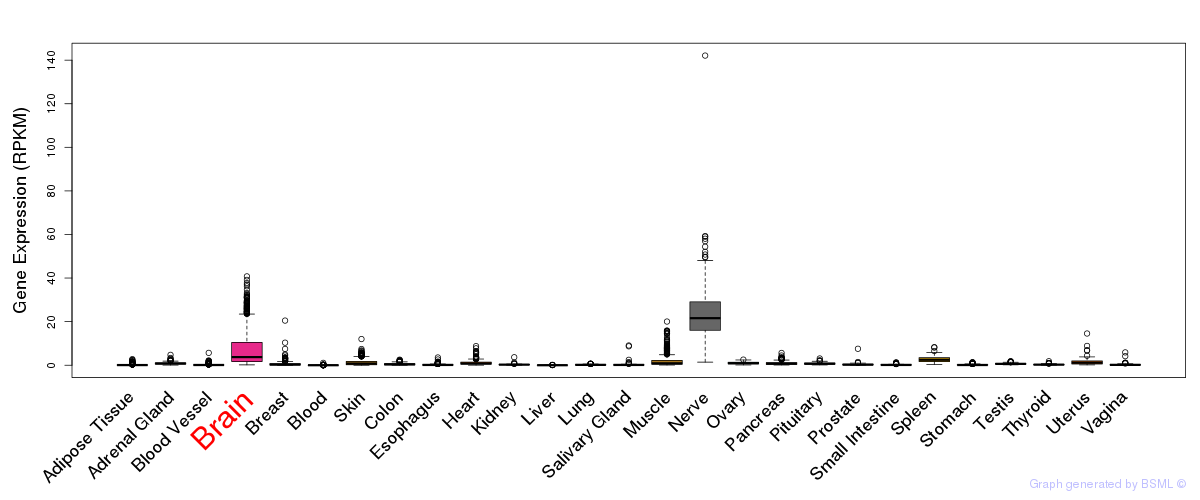
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NUP160 | 0.95 | 0.92 |
| FANCC | 0.95 | 0.91 |
| SMAD4 | 0.95 | 0.92 |
| SNRNP200 | 0.94 | 0.93 |
| PATZ1 | 0.94 | 0.91 |
| VEZF1 | 0.94 | 0.86 |
| KDM5B | 0.93 | 0.93 |
| NUP188 | 0.93 | 0.89 |
| PRRC1 | 0.93 | 0.93 |
| ZBTB5 | 0.93 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.67 | -0.79 |
| HLA-F | -0.67 | -0.80 |
| FBXO2 | -0.67 | -0.67 |
| AIFM3 | -0.65 | -0.76 |
| PTH1R | -0.64 | -0.72 |
| LHPP | -0.64 | -0.60 |
| ALDOC | -0.63 | -0.69 |
| TNFSF12 | -0.63 | -0.66 |
| CA4 | -0.63 | -0.74 |
| ASPHD1 | -0.63 | -0.57 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOSAMINOGLYCAN BIOSYNTHESIS HEPARAN SULFATE | 26 | 16 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| MANTOVANI VIRAL GPCR SIGNALING DN | 49 | 37 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS UP | 88 | 47 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION UP | 74 | 40 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| CHO NR4A1 TARGETS | 33 | 22 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K4ME3 AND H3K27ME3 | 38 | 34 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K4ME3 AND H327ME3 | 126 | 83 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 AND H3K27ME3 | 137 | 85 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |