| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS
| 97 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION
| 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS
| 140 | 100 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY
| 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN2 PATHWAY
| 29 | 18 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP
| 351 | 230 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP
| 294 | 178 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP
| 579 | 346 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP
| 1382 | 904 | All SZGR 2.0 genes in this pathway |
| SMIRNOV CIRCULATING ENDOTHELIOCYTES IN CANCER UP
| 158 | 103 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP
| 380 | 236 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN
| 187 | 115 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN
| 226 | 132 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN
| 1375 | 806 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP
| 443 | 294 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN
| 712 | 443 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS
| 183 | 115 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS
| 882 | 572 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP
| 165 | 100 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY UP
| 61 | 34 | All SZGR 2.0 genes in this pathway |
| ADDYA ERYTHROID DIFFERENTIATION BY HEMIN
| 73 | 47 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP
| 114 | 84 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN
| 128 | 90 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 6M
| 74 | 47 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY UP
| 45 | 29 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 3M
| 59 | 36 | All SZGR 2.0 genes in this pathway |
| CHEN ETV5 TARGETS SERTOLI
| 20 | 10 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN
| 245 | 144 | All SZGR 2.0 genes in this pathway |
| VISALA RESPONSE TO HEAT SHOCK AND AGING DN
| 14 | 12 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP
| 253 | 147 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P2
| 79 | 55 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED
| 1229 | 713 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN
| 940 | 425 | All SZGR 2.0 genes in this pathway |
| MARTINELLI IMMATURE NEUTROPHIL DN
| 13 | 9 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE
| 89 | 56 | All SZGR 2.0 genes in this pathway |
| HOFFMANN SMALL PRE BII TO IMMATURE B LYMPHOCYTE DN
| 50 | 33 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK
| 1210 | 725 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP
| 288 | 168 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1
| 237 | 159 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP
| 307 | 182 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL
| 216 | 130 | All SZGR 2.0 genes in this pathway |
| DEMAGALHAES AGING UP
| 55 | 39 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP
| 387 | 225 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP
| 217 | 131 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN
| 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| KIM GLIS2 TARGETS UP
| 84 | 61 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP
| 149 | 96 | All SZGR 2.0 genes in this pathway |
| ALTEMEIER RESPONSE TO LPS WITH MECHANICAL VENTILATION
| 128 | 81 | All SZGR 2.0 genes in this pathway |
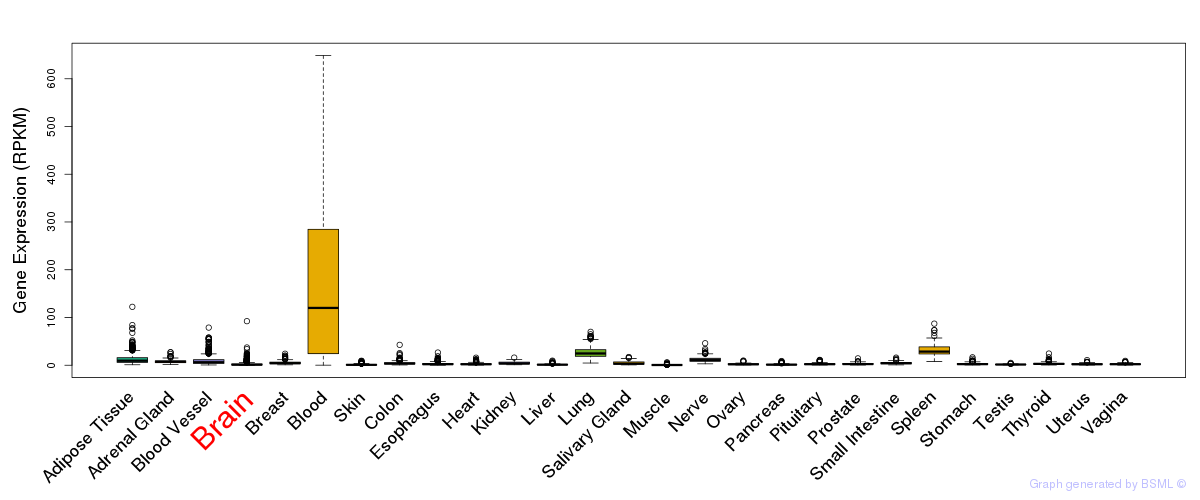
 eQTL annotation
eQTL annotation