Gene Page: CLCA4
Summary ?
| GeneID | 22802 |
| Symbol | CLCA4 |
| Synonyms | CaCC|CaCC2 |
| Description | chloride channel accessory 4 |
| Reference | MIM:616857|HGNC:HGNC:2018|Ensembl:ENSG00000016602|HPRD:16719|Vega:OTTHUMG00000010260 |
| Gene type | protein-coding |
| Map location | 1p22.3 |
| Pascal p-value | 0.816 |
| Sherlock p-value | 0.105 |
| Fetal beta | -2.116 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10690677 | 1 | 87019175 | CLCA4 | 3.53E-5 | 0.578 | 0.02 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| snp_a-2241490 | 0 | CLCA4 | 22802 | 0.16 | trans | |||
| snp_a-4293886 | 0 | CLCA4 | 22802 | 0.19 | trans | |||
| rs17152736 | chr7 | 79290748 | CLCA4 | 22802 | 0 | trans | ||
| rs12575741 | chr11 | 35969675 | CLCA4 | 22802 | 0.16 | trans | ||
| rs7888140 | chrX | 19825933 | CLCA4 | 22802 | 0.19 | trans | ||
| rs7876405 | chrX | 136170379 | CLCA4 | 22802 | 0.02 | trans |
Section II. Transcriptome annotation
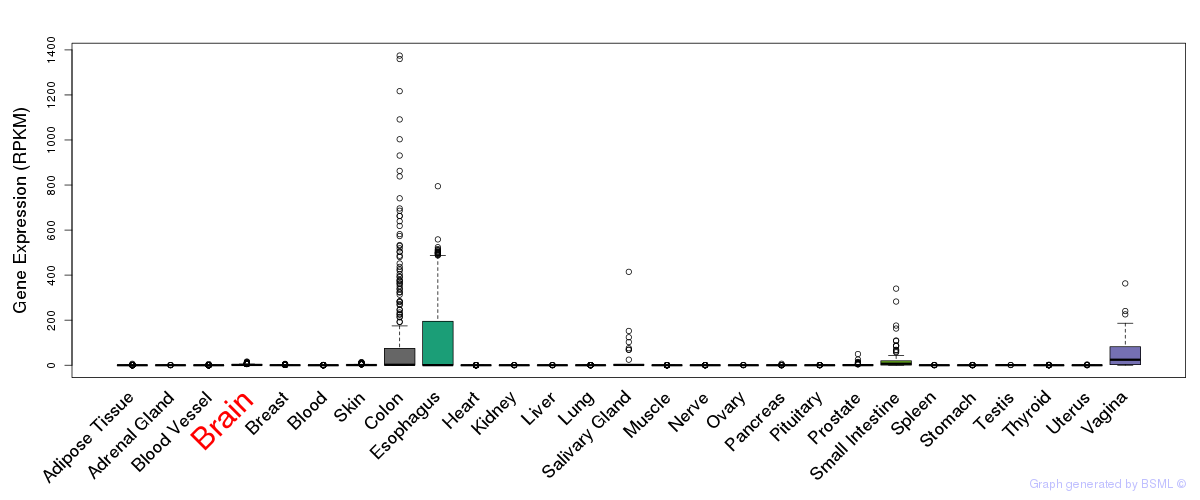
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GOPC | 0.92 | 0.93 |
| CAMSAP1L1 | 0.92 | 0.94 |
| RAB14 | 0.91 | 0.93 |
| RBBP5 | 0.91 | 0.93 |
| C4orf41 | 0.91 | 0.92 |
| KIAA1219 | 0.91 | 0.93 |
| NUP50 | 0.91 | 0.93 |
| UBQLN1 | 0.91 | 0.92 |
| ARCN1 | 0.91 | 0.92 |
| FBXO11 | 0.91 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.75 | -0.78 |
| MT-CO2 | -0.73 | -0.79 |
| AF347015.31 | -0.73 | -0.78 |
| HIGD1B | -0.72 | -0.78 |
| TLCD1 | -0.71 | -0.74 |
| MT-CYB | -0.70 | -0.75 |
| AF347015.33 | -0.70 | -0.73 |
| AF347015.8 | -0.70 | -0.77 |
| ENHO | -0.69 | -0.79 |
| HSD17B14 | -0.69 | -0.72 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG OLFACTORY TRANSDUCTION | 389 | 85 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY UP | 109 | 69 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER E | 89 | 44 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA NEURAL | 129 | 85 | All SZGR 2.0 genes in this pathway |
| BOSCO EPITHELIAL DIFFERENTIATION MODULE | 69 | 31 | All SZGR 2.0 genes in this pathway |