Gene Page: ZNF507
Summary ?
| GeneID | 22847 |
| Symbol | ZNF507 |
| Synonyms | - |
| Description | zinc finger protein 507 |
| Reference | HGNC:HGNC:23783|Ensembl:ENSG00000168813|HPRD:11722|Vega:OTTHUMG00000180220 |
| Gene type | protein-coding |
| Map location | 19q13.11 |
| Pascal p-value | 0.861 |
| Fetal beta | 0.258 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
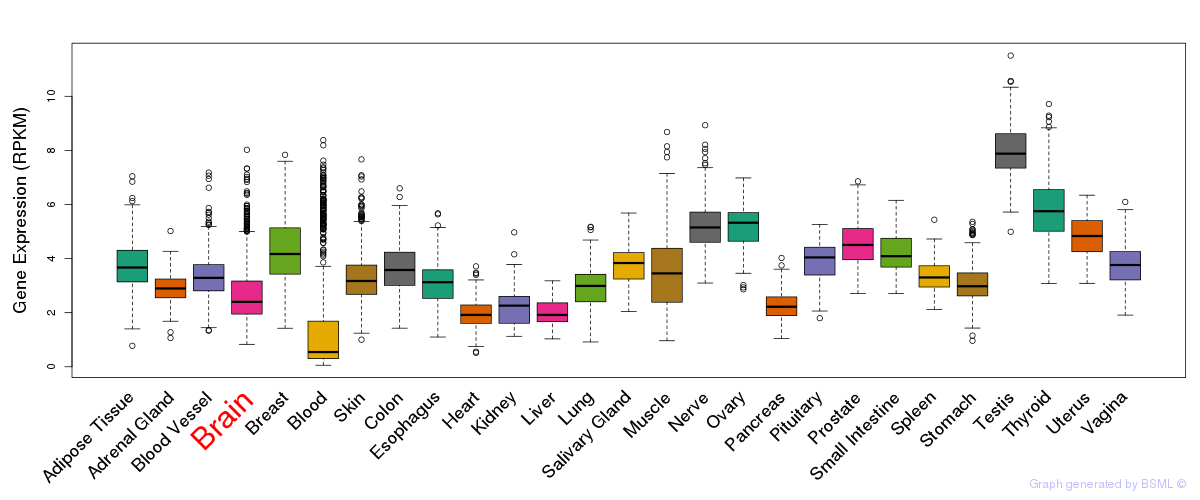
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KCNB1 | 0.95 | 0.88 |
| KCNH1 | 0.94 | 0.79 |
| SCN8A | 0.94 | 0.85 |
| STXBP5L | 0.93 | 0.85 |
| PLCB1 | 0.93 | 0.86 |
| KCNMA1 | 0.93 | 0.88 |
| CREG2 | 0.92 | 0.73 |
| GABRB2 | 0.92 | 0.84 |
| SLC7A14 | 0.92 | 0.84 |
| IQSEC1 | 0.92 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.50 | -0.62 |
| AC006276.2 | -0.46 | -0.48 |
| C9orf46 | -0.44 | -0.49 |
| RPLP1 | -0.44 | -0.51 |
| RPL36 | -0.43 | -0.54 |
| RPL35 | -0.43 | -0.54 |
| RPL23A | -0.43 | -0.49 |
| HEBP2 | -0.42 | -0.58 |
| RPS18 | -0.42 | -0.49 |
| NME4 | -0.42 | -0.50 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY 4NQO IN WS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY GAMMA IN OLD | 31 | 19 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY GAMMA IN WS | 33 | 22 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY UV IN OLD | 25 | 18 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY UV IN WS | 12 | 9 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE DN | 21 | 12 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D UP | 89 | 62 | All SZGR 2.0 genes in this pathway |