Gene Page: PLEKHA6
Summary ?
| GeneID | 22874 |
| Symbol | PLEKHA6 |
| Synonyms | PEPP-3|PEPP3 |
| Description | pleckstrin homology domain containing A6 |
| Reference | MIM:607771|HGNC:HGNC:17053|Ensembl:ENSG00000143850|HPRD:09685|Vega:OTTHUMG00000036057 |
| Gene type | protein-coding |
| Map location | 1q32.1 |
| Sherlock p-value | 0.575 |
| Fetal beta | -0.768 |
| DMG | 1 (# studies) |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PLEKHA6 | chr1 | 204236636 | C | T | NM_014935 | p.83V>I | missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24537993 | 1 | 204275926 | PLEKHA6 | 1.852E-4 | 0.347 | 0.034 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
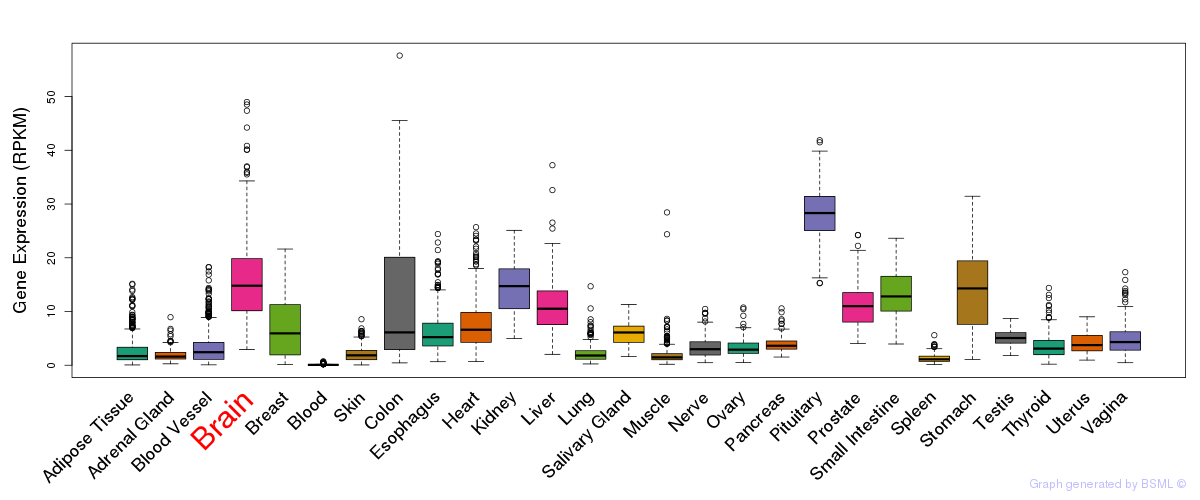
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF783 | 0.92 | 0.93 |
| QRICH2 | 0.91 | 0.91 |
| FBN3 | 0.90 | 0.85 |
| NINL | 0.90 | 0.89 |
| ANO8 | 0.90 | 0.91 |
| ADAMTS10 | 0.89 | 0.88 |
| AC010536.2 | 0.89 | 0.89 |
| GIGYF1 | 0.89 | 0.90 |
| BAHCC1 | 0.88 | 0.91 |
| BTBD12 | 0.88 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.61 | -0.83 |
| C5orf53 | -0.60 | -0.72 |
| S100B | -0.60 | -0.78 |
| HLA-F | -0.60 | -0.67 |
| B2M | -0.60 | -0.77 |
| AF347015.27 | -0.60 | -0.81 |
| ALDOC | -0.60 | -0.64 |
| ABCG2 | -0.59 | -0.60 |
| COPZ2 | -0.59 | -0.76 |
| AIFM3 | -0.59 | -0.67 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL DN | 114 | 58 | All SZGR 2.0 genes in this pathway |
| TCGA GLIOBLASTOMA COPY NUMBER UP | 75 | 36 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER ERBB2 UP | 147 | 83 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS DN | 23 | 16 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 LCP WITH H3K4ME3 | 162 | 80 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS DN | 115 | 73 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| GHANDHI DIRECT IRRADIATION DN | 33 | 23 | All SZGR 2.0 genes in this pathway |