Gene Page: CUL9
Summary ?
| GeneID | 23113 |
| Symbol | CUL9 |
| Synonyms | H7AP1|PARC |
| Description | cullin 9 |
| Reference | MIM:607489|HGNC:HGNC:15982|Ensembl:ENSG00000112659|HPRD:06317|Vega:OTTHUMG00000014723 |
| Gene type | protein-coding |
| Map location | 6p21.1 |
| Pascal p-value | 3.244E-5 |
| Sherlock p-value | 0.609 |
| Fetal beta | -0.577 |
| eGene | Caudate basal ganglia Cortex |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
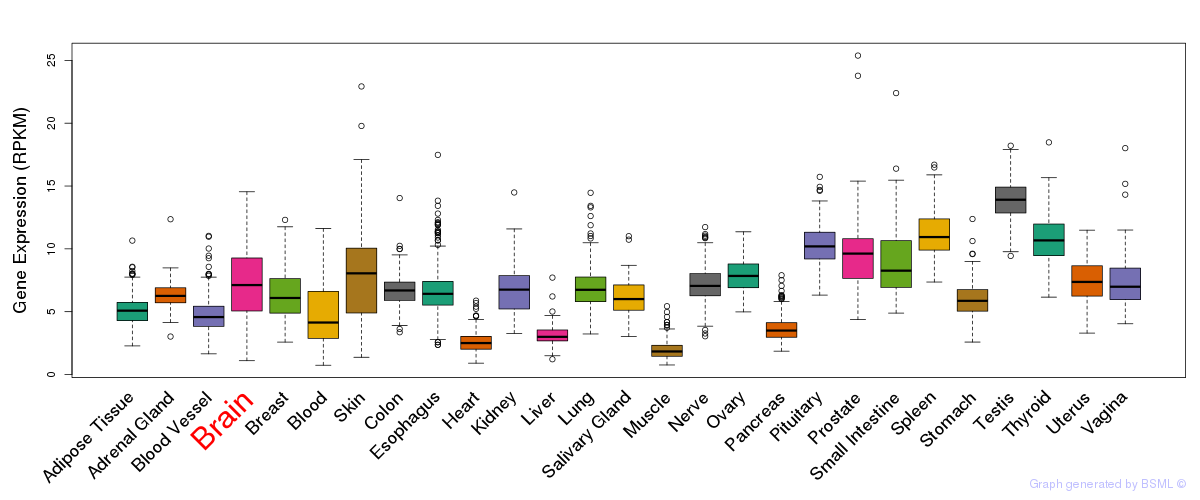
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MGA | 0.97 | 0.97 |
| BIRC6 | 0.97 | 0.97 |
| C10orf18 | 0.96 | 0.96 |
| NSD1 | 0.96 | 0.95 |
| UBR5 | 0.96 | 0.96 |
| TUG1 | 0.95 | 0.96 |
| MED1 | 0.95 | 0.96 |
| SBF2 | 0.95 | 0.95 |
| ZNF445 | 0.95 | 0.94 |
| CLASP1 | 0.95 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.71 | -0.83 |
| MT-CO2 | -0.70 | -0.84 |
| IFI27 | -0.70 | -0.83 |
| FXYD1 | -0.70 | -0.81 |
| C5orf53 | -0.69 | -0.72 |
| HIGD1B | -0.68 | -0.84 |
| AF347015.27 | -0.68 | -0.79 |
| S100B | -0.68 | -0.77 |
| AF347015.33 | -0.67 | -0.78 |
| HLA-F | -0.67 | -0.70 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 18230339 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0031625 | ubiquitin protein ligase binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006511 | ubiquitin-dependent protein catabolic process | IEA | - | |
| GO:0030071 | regulation of mitotic metaphase/anaphase transition | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005680 | anaphase-promoting complex | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0031461 | cullin-RING ubiquitin ligase complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN | 175 | 82 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| SCHWAB TARGETS OF BMYB POLYMORPHIC VARIANTS DN | 17 | 12 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY TARGETS OF MUTATED TP53 DN | 50 | 24 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 UP | 170 | 97 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP | 140 | 81 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 7 UP | 118 | 68 | All SZGR 2.0 genes in this pathway |