Gene Page: FMOD
Summary ?
| GeneID | 2331 |
| Symbol | FMOD |
| Synonyms | FM|SLRR2E |
| Description | fibromodulin |
| Reference | MIM:600245|HGNC:HGNC:3774|Ensembl:ENSG00000122176|HPRD:02591|Vega:OTTHUMG00000035910 |
| Gene type | protein-coding |
| Map location | 1q32 |
| eGene | Frontal Cortex BA9 Nucleus accumbens basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17131472 | chr1 | 91962966 | FMOD | 2331 | 0.02 | trans | ||
| rs13388853 | 0 | FMOD | 2331 | 0.12 | trans | |||
| rs1038697 | chr2 | 18537074 | FMOD | 2331 | 0.06 | trans | ||
| rs17015803 | chr2 | 120191430 | FMOD | 2331 | 0.01 | trans | ||
| rs1691463 | chr3 | 84914366 | FMOD | 2331 | 0.1 | trans | ||
| rs2922180 | chr3 | 125280472 | FMOD | 2331 | 0 | trans | ||
| rs2971271 | chr3 | 125450402 | FMOD | 2331 | 0.17 | trans | ||
| rs2976809 | chr3 | 125451738 | FMOD | 2331 | 6.179E-4 | trans | ||
| rs3796892 | chr4 | 111404774 | FMOD | 2331 | 0.18 | trans | ||
| rs313946 | chr4 | 113975904 | FMOD | 2331 | 0.01 | trans | ||
| rs657141 | chr4 | 113981041 | FMOD | 2331 | 0.02 | trans | ||
| rs10050805 | chr5 | 30805352 | FMOD | 2331 | 0.11 | trans | ||
| rs16894149 | chr5 | 61013724 | FMOD | 2331 | 0.1 | trans | ||
| rs6449550 | chr5 | 61034312 | FMOD | 2331 | 0.08 | trans | ||
| rs6874540 | chr5 | 149911669 | FMOD | 2331 | 0.04 | trans | ||
| rs2273232 | chr5 | 149912526 | FMOD | 2331 | 0.04 | trans | ||
| rs17056435 | chr5 | 158453782 | FMOD | 2331 | 0.14 | trans | ||
| rs2294262 | chr6 | 16108966 | FMOD | 2331 | 0.19 | trans | ||
| rs4236095 | chr6 | 47368686 | FMOD | 2331 | 0.09 | trans | ||
| rs6455226 | chr6 | 67930295 | FMOD | 2331 | 0 | trans | ||
| rs7841407 | chr8 | 9243427 | FMOD | 2331 | 0.06 | trans | ||
| rs9423711 | chr10 | 2574802 | FMOD | 2331 | 0.15 | trans | ||
| rs7081666 | chr10 | 47673800 | FMOD | 2331 | 0.1 | trans | ||
| rs11820030 | chr11 | 76133564 | FMOD | 2331 | 0.19 | trans | ||
| rs17134872 | chr11 | 76139845 | FMOD | 2331 | 0.06 | trans | ||
| rs10751255 | 0 | FMOD | 2331 | 0.19 | trans | |||
| rs11236774 | chr11 | 76234952 | FMOD | 2331 | 0.06 | trans | ||
| rs10879027 | chr12 | 70237156 | FMOD | 2331 | 0.06 | trans | ||
| rs11112163 | chr12 | 105072508 | FMOD | 2331 | 0.03 | trans | ||
| rs9529974 | chr13 | 72821935 | FMOD | 2331 | 0 | trans | ||
| rs17073665 | chr13 | 81541961 | FMOD | 2331 | 0.14 | trans | ||
| rs17657985 | chr13 | 100499573 | FMOD | 2331 | 0.17 | trans | ||
| rs2093759 | chr14 | 97171024 | FMOD | 2331 | 0.04 | trans | ||
| rs17244419 | chr14 | 97171074 | FMOD | 2331 | 0.01 | trans | ||
| rs11873703 | chr18 | 61448702 | FMOD | 2331 | 2.012E-4 | trans | ||
| rs6095741 | chr20 | 48666589 | FMOD | 2331 | 4.443E-6 | trans | ||
| rs5991662 | chrX | 43335796 | FMOD | 2331 | 0.17 | trans | ||
| rs1545676 | chrX | 93530472 | FMOD | 2331 | 0.07 | trans | ||
| rs3761581 | chrX | 128789720 | FMOD | 2331 | 0.16 | trans | ||
| rs242163 | chrX | 132311662 | FMOD | 2331 | 0.14 | trans |
Section II. Transcriptome annotation
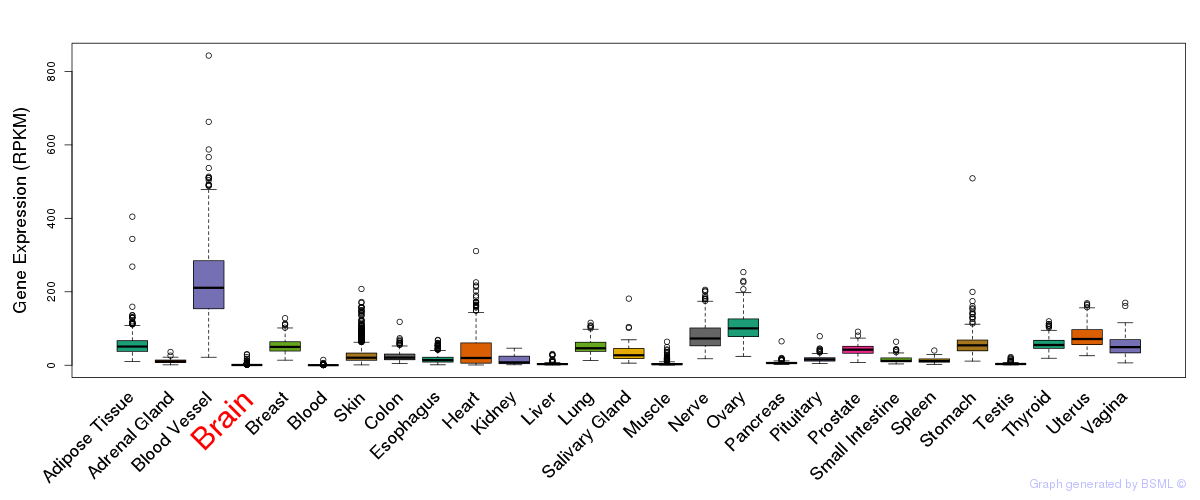
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME KERATAN SULFATE BIOSYNTHESIS | 26 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME KERATAN SULFATE KERATIN METABOLISM | 30 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME KERATAN SULFATE DEGRADATION | 11 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSAMINOGLYCAN METABOLISM | 111 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| PAPASPYRIDONOS UNSTABLE ATEROSCLEROTIC PLAQUE DN | 43 | 29 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL UP | 121 | 72 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| XU HGF TARGETS REPRESSED BY AKT1 DN | 95 | 58 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA DN | 28 | 21 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI UP | 92 | 58 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT DN | 80 | 56 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 UP | 84 | 55 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| LY AGING MIDDLE UP | 14 | 11 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR UP | 101 | 69 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER BRCA1 DN | 44 | 28 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| ELLWOOD MYC TARGETS DN | 40 | 27 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER WITH EPCAM UP | 53 | 25 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ CHRONIC LYMPHOCYTIC LEUKEMIA DN | 56 | 39 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| LY AGING OLD UP | 7 | 5 | All SZGR 2.0 genes in this pathway |
| NOUSHMEHR GBM SILENCED BY METHYLATION | 50 | 32 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 5 DN | 50 | 31 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA PROTEOGLYCANS | 35 | 23 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |