Gene Page: UBR4
Summary ?
| GeneID | 23352 |
| Symbol | UBR4 |
| Synonyms | RBAF600|ZUBR1|p600 |
| Description | ubiquitin protein ligase E3 component n-recognin 4 |
| Reference | MIM:609890|HGNC:HGNC:30313|Ensembl:ENSG00000127481|HPRD:10184|Vega:OTTHUMG00000002498 |
| Gene type | protein-coding |
| Map location | 1p36.13 |
| Pascal p-value | 0.424 |
| Sherlock p-value | 0.998 |
| Fetal beta | 0.67 |
| eGene | Myers' cis & trans Meta |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11261013 | chr1 | 18422971 | UBR4 | 23352 | 0.17 | cis | ||
| rs999974 | chr1 | 19793983 | UBR4 | 23352 | 0.09 | cis |
Section II. Transcriptome annotation
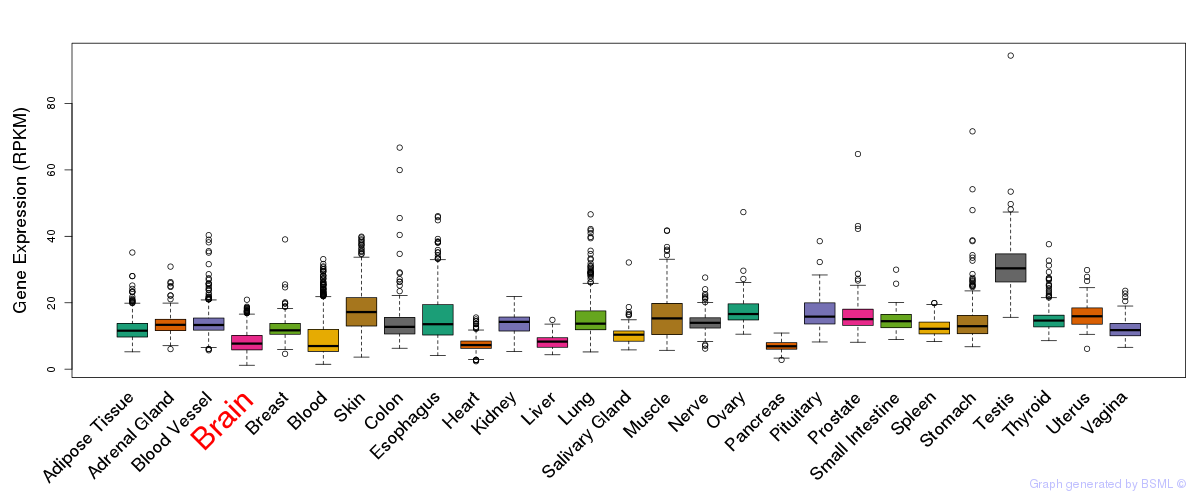
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MTA2 | 0.98 | 0.94 |
| NUP62 | 0.97 | 0.92 |
| PHF2 | 0.96 | 0.95 |
| BRD1 | 0.96 | 0.93 |
| CSNK1E | 0.96 | 0.90 |
| HNRNPA0 | 0.96 | 0.91 |
| ZSCAN2 | 0.96 | 0.90 |
| GEMIN4 | 0.96 | 0.90 |
| KDM4A | 0.96 | 0.89 |
| TRIM28 | 0.96 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.75 | -0.78 |
| HLA-F | -0.72 | -0.71 |
| AIFM3 | -0.70 | -0.67 |
| AF347015.27 | -0.69 | -0.84 |
| FBXO2 | -0.69 | -0.59 |
| AF347015.31 | -0.69 | -0.84 |
| S100B | -0.69 | -0.77 |
| PTH1R | -0.69 | -0.67 |
| ALDOC | -0.68 | -0.61 |
| MT-CO2 | -0.68 | -0.86 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER HIGH RECURRENCE | 49 | 31 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC UP | 33 | 20 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC 24HR 5 DN | 59 | 35 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS DN | 86 | 67 | All SZGR 2.0 genes in this pathway |
| MARIADASON REGULATED BY HISTONE ACETYLATION UP | 83 | 49 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |