Gene Page: VPS8
Summary ?
| GeneID | 23355 |
| Symbol | VPS8 |
| Synonyms | KIAA0804 |
| Description | VPS8, CORVET complex subunit |
| Reference | HGNC:HGNC:29122|Ensembl:ENSG00000156931|HPRD:11111|Vega:OTTHUMG00000156707 |
| Gene type | protein-coding |
| Map location | 3q27.2 |
| Pascal p-value | 0.249 |
| Sherlock p-value | 3.097E-5 |
| Fetal beta | -0.033 |
| eGene | Caudate basal ganglia Cerebellum Frontal Cortex BA9 Nucleus accumbens basal ganglia Putamen basal ganglia |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs62286935 | 3 | 184555231 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 25300 | gtex_brain_putamen_basal |
| rs62286936 | 3 | 184556483 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 26552 | gtex_brain_putamen_basal |
| rs61741194 | 3 | 184587274 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 57343 | gtex_brain_putamen_basal |
| rs80051460 | 3 | 184587926 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 57995 | gtex_brain_putamen_basal |
| rs201291790 | 3 | 184620289 | VPS8 | ENSG00000156931.11 | 3.785E-6 | 0.02 | 90358 | gtex_brain_putamen_basal |
| rs62286955 | 3 | 184621202 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 91271 | gtex_brain_putamen_basal |
| rs62286956 | 3 | 184622725 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 92794 | gtex_brain_putamen_basal |
| rs62286959 | 3 | 184638094 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 108163 | gtex_brain_putamen_basal |
| rs112843941 | 3 | 184645378 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 115447 | gtex_brain_putamen_basal |
| rs62289479 | 3 | 184711397 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 181466 | gtex_brain_putamen_basal |
| rs145841670 | 3 | 184718460 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 188529 | gtex_brain_putamen_basal |
| rs74823827 | 3 | 184727815 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 197884 | gtex_brain_putamen_basal |
| rs62289499 | 3 | 184730390 | VPS8 | ENSG00000156931.11 | 1.339E-6 | 0.02 | 200459 | gtex_brain_putamen_basal |
| rs116713356 | 3 | 184736990 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 207059 | gtex_brain_putamen_basal |
| rs62289500 | 3 | 184737550 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 207619 | gtex_brain_putamen_basal |
| rs62289501 | 3 | 184737911 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 207980 | gtex_brain_putamen_basal |
| rs80288863 | 3 | 184760314 | VPS8 | ENSG00000156931.11 | 1.598E-6 | 0.02 | 230383 | gtex_brain_putamen_basal |
| rs62287919 | 3 | 184784607 | VPS8 | ENSG00000156931.11 | 1.6E-6 | 0.02 | 254676 | gtex_brain_putamen_basal |
| rs57628070 | 3 | 184788052 | VPS8 | ENSG00000156931.11 | 1.571E-6 | 0.02 | 258121 | gtex_brain_putamen_basal |
| rs77142485 | 3 | 184788130 | VPS8 | ENSG00000156931.11 | 1.603E-6 | 0.02 | 258199 | gtex_brain_putamen_basal |
| rs62287921 | 3 | 184790415 | VPS8 | ENSG00000156931.11 | 1.578E-6 | 0.02 | 260484 | gtex_brain_putamen_basal |
| rs62287922 | 3 | 184790949 | VPS8 | ENSG00000156931.11 | 1.554E-6 | 0.02 | 261018 | gtex_brain_putamen_basal |
| rs62287923 | 3 | 184792190 | VPS8 | ENSG00000156931.11 | 1.5E-6 | 0.02 | 262259 | gtex_brain_putamen_basal |
| rs62287925 | 3 | 184792815 | VPS8 | ENSG00000156931.11 | 1.489E-6 | 0.02 | 262884 | gtex_brain_putamen_basal |
| rs62287929 | 3 | 184795079 | VPS8 | ENSG00000156931.11 | 3.019E-6 | 0.02 | 265148 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
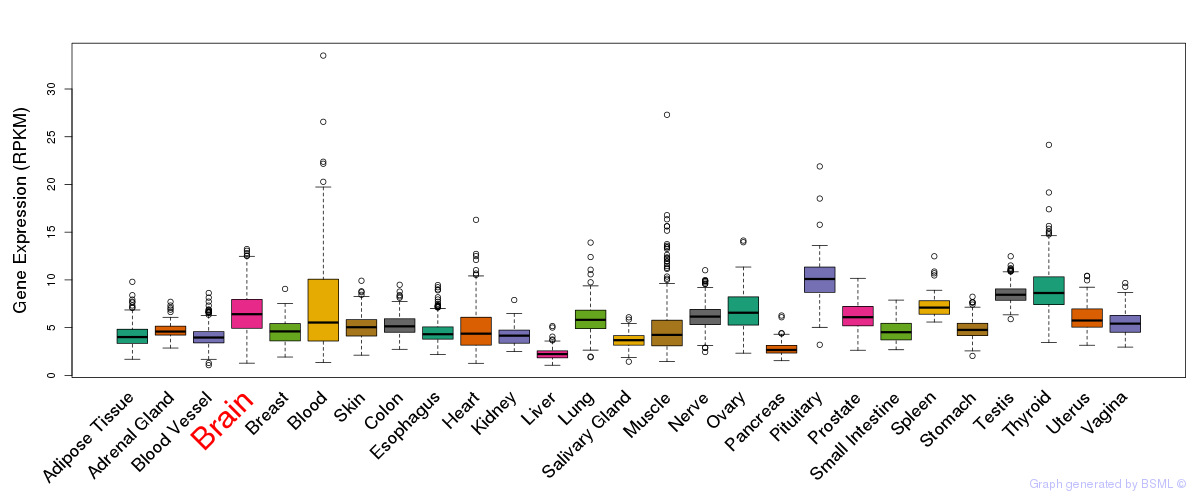
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU SOX4 TARGETS UP | 137 | 94 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D DN | 143 | 83 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER EXPRESSION BY COPY NUMBER | 100 | 62 | All SZGR 2.0 genes in this pathway |
| NOUZOVA METHYLATED IN APL | 68 | 39 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |