Gene Page: QPRT
Summary ?
| GeneID | 23475 |
| Symbol | QPRT |
| Synonyms | HEL-S-90n|QPRTase |
| Description | quinolinate phosphoribosyltransferase |
| Reference | MIM:606248|HGNC:HGNC:9755|HPRD:06950| |
| Gene type | protein-coding |
| Map location | 16p11.2 |
| Pascal p-value | 0.361 |
| Fetal beta | -0.895 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Cortex Frontal Cortex BA9 Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05000199 | 16 | 29701173 | QPRT | 4.716E-4 | 0.259 | 0.046 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | QPRT | 23475 | 0.07 | trans | ||
| rs62059023 | 16 | 29681707 | QPRT | ENSG00000103485.13 | 1.346E-6 | 0 | -8622 | gtex_brain_ba24 |
| rs62059024 | 16 | 29681734 | QPRT | ENSG00000103485.13 | 1.342E-6 | 0 | -8595 | gtex_brain_ba24 |
| rs12447415 | 16 | 29682945 | QPRT | ENSG00000103485.13 | 2.932E-7 | 0 | -7384 | gtex_brain_ba24 |
| rs17842268 | 16 | 29684180 | QPRT | ENSG00000103485.13 | 3.045E-7 | 0 | -6149 | gtex_brain_ba24 |
| rs8058049 | 16 | 29684572 | QPRT | ENSG00000103485.13 | 3.053E-7 | 0 | -5757 | gtex_brain_ba24 |
| rs9940440 | 16 | 29684788 | QPRT | ENSG00000103485.13 | 2.932E-7 | 0 | -5541 | gtex_brain_ba24 |
| rs79706398 | 16 | 29688045 | QPRT | ENSG00000103485.13 | 4.66E-7 | 0 | -2284 | gtex_brain_ba24 |
| rs9933310 | 16 | 29690904 | QPRT | ENSG00000103485.13 | 4.876E-7 | 0 | 575 | gtex_brain_ba24 |
| rs62059025 | 16 | 29692277 | QPRT | ENSG00000103485.13 | 1.046E-6 | 0 | 1948 | gtex_brain_ba24 |
| rs148320072 | 16 | 29694389 | QPRT | ENSG00000103485.13 | 2.029E-7 | 0 | 4060 | gtex_brain_ba24 |
| rs141987948 | 16 | 29694390 | QPRT | ENSG00000103485.13 | 2.028E-7 | 0 | 4061 | gtex_brain_ba24 |
| rs145634669 | 16 | 29694391 | QPRT | ENSG00000103485.13 | 2.028E-7 | 0 | 4062 | gtex_brain_ba24 |
| rs11644909 | 16 | 29694414 | QPRT | ENSG00000103485.13 | 1.824E-7 | 0 | 4085 | gtex_brain_ba24 |
Section II. Transcriptome annotation
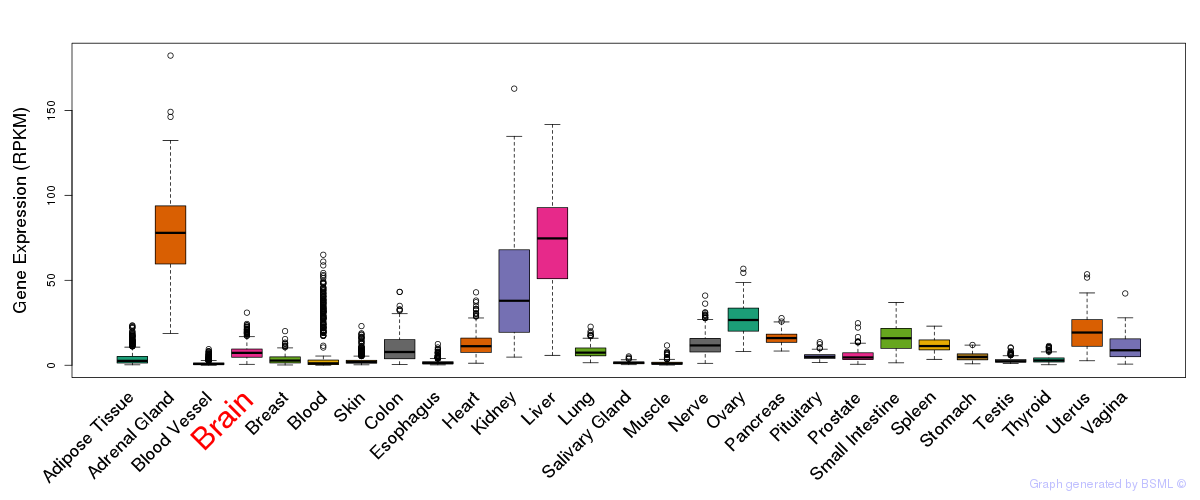
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity | EXP | 9473669 | |
| GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity | IDA | 3409840 |9473669 |17868694 | |
| GO:0016757 | transferase activity, transferring glycosyl groups | IEA | - | |
| GO:0042803 | protein homodimerization activity | IDA | 17868694 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0009435 | NAD biosynthetic process | IEA | - | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0034213 | quinolinate catabolic process | IDA | 9473669 | |
| GO:0051259 | protein oligomerization | IDA | 17868694 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 9473669 | |
| GO:0005575 | cellular_component | ND | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NICOTINATE AND NICOTINAMIDE METABOLISM | 24 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF VITAMINS AND COFACTORS | 51 | 36 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 UP | 151 | 84 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 3D UP | 182 | 110 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINED IN MONOCYTE UP | 21 | 15 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINDED IN ERYTHROCYTE UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL UP | 146 | 99 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| KOINUMA COLON CANCER MSI DN | 8 | 5 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION UP | 126 | 78 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW DN | 165 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH DN | 180 | 110 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA DN | 181 | 107 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| WATTEL AUTONOMOUS THYROID ADENOMA UP | 73 | 47 | All SZGR 2.0 genes in this pathway |
| ALCALA APOPTOSIS | 88 | 60 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION UP | 140 | 83 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER ERBB2 UP | 147 | 83 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 3 | 33 | 12 | All SZGR 2.0 genes in this pathway |
| VALK AML WITH FLT3 ITD | 40 | 22 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K27ME3 | 206 | 108 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| VANLOO SP3 TARGETS DN | 89 | 47 | All SZGR 2.0 genes in this pathway |