| KEGG GALACTOSE METABOLISM
| 26 | 22 | All SZGR 2.0 genes in this pathway |
| KEGG AMINO SUGAR AND NUCLEOTIDE SUGAR METABOLISM
| 44 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES
| 247 | 154 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 DN
| 153 | 100 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN
| 234 | 147 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION DN
| 84 | 54 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN
| 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN
| 770 | 415 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN
| 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP
| 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP
| 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP
| 146 | 104 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD UP
| 64 | 39 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP
| 176 | 111 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP
| 227 | 137 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON
| 335 | 181 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION
| 171 | 102 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION UP
| 71 | 51 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION
| 475 | 313 | All SZGR 2.0 genes in this pathway |
| BRUNO HEMATOPOIESIS
| 66 | 48 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN
| 86 | 66 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 24HR
| 43 | 30 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS INTERMEDIATE PROGENITOR
| 149 | 84 | All SZGR 2.0 genes in this pathway |
| SU LIVER
| 55 | 32 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP
| 428 | 266 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN
| 287 | 208 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN
| 96 | 71 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN
| 141 | 92 | All SZGR 2.0 genes in this pathway |
| HANN RESISTANCE TO BCL2 INHIBITOR DN
| 48 | 31 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING
| 510 | 309 | All SZGR 2.0 genes in this pathway |
| HOFFMANN IMMATURE TO MATURE B LYMPHOCYTE DN
| 50 | 36 | All SZGR 2.0 genes in this pathway |
| CROONQUIST IL6 DEPRIVATION DN
| 98 | 69 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14
| 143 | 86 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 3 UP
| 89 | 50 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP
| 163 | 111 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS UP
| 135 | 96 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS DN
| 53 | 29 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY
| 1725 | 838 | All SZGR 2.0 genes in this pathway |
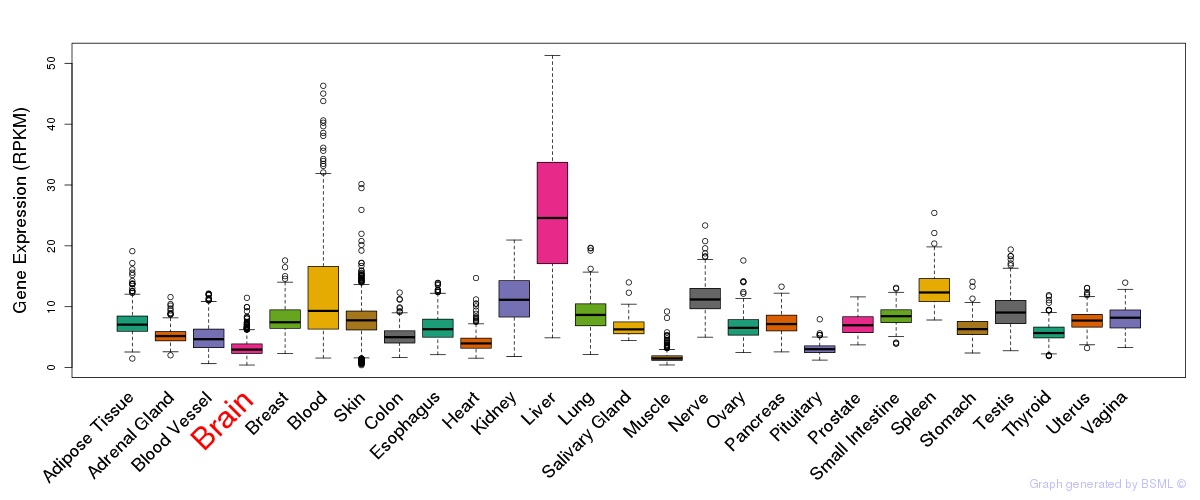
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation