Gene Page: ZDHHC5
Summary ?
| GeneID | 25921 |
| Symbol | ZDHHC5 |
| Synonyms | DHHC5|ZNF375 |
| Description | zinc finger DHHC-type containing 5 |
| Reference | MIM:614586|HGNC:HGNC:18472|Ensembl:ENSG00000156599|HPRD:15720|Vega:OTTHUMG00000167198 |
| Gene type | protein-coding |
| Map location | 11q12.1 |
| Pascal p-value | 2.32E-7 |
| Sherlock p-value | 0.482 |
| TADA p-value | 0.002 |
| Fetal beta | -0.663 |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.441 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ZDHHC5 | chr11 | 57466850 | G | T | NM_015457 | p.648E>* | nonsense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10834670 | chr11 | 25404015 | ZDHHC5 | 25921 | 0.11 | trans | ||
| rs10834671 | chr11 | 25404269 | ZDHHC5 | 25921 | 0.07 | trans |
Section II. Transcriptome annotation
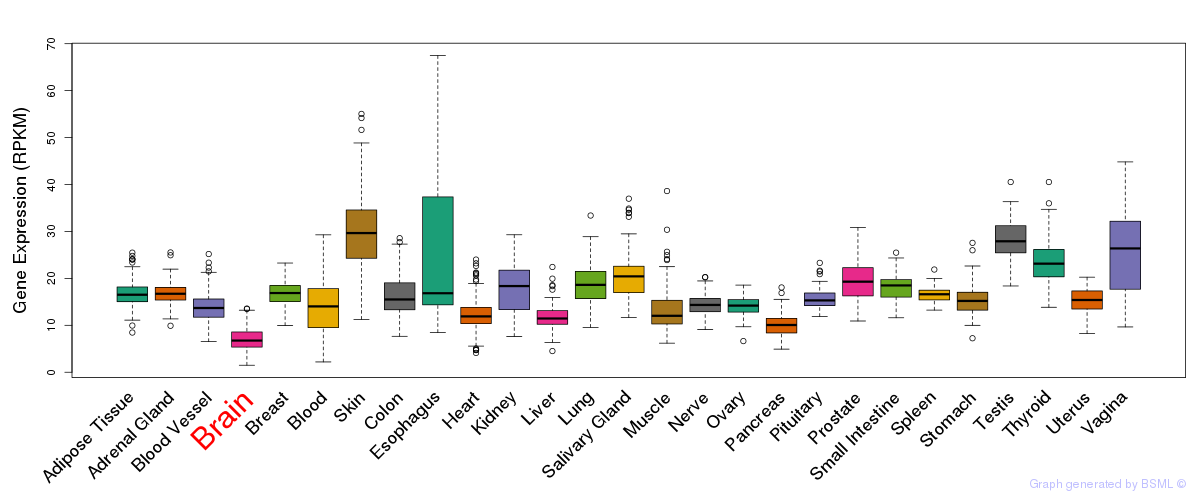
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EXOSC8 | 0.77 | 0.78 |
| SNRPF | 0.77 | 0.80 |
| DYNLT1 | 0.77 | 0.79 |
| RFC4 | 0.76 | 0.77 |
| LSM8 | 0.75 | 0.72 |
| PDSS1 | 0.75 | 0.74 |
| C14orf109 | 0.74 | 0.73 |
| AC106037.2 | 0.74 | 0.74 |
| KDELC1 | 0.74 | 0.71 |
| CBY1 | 0.74 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.57 | -0.64 |
| FBXO2 | -0.55 | -0.58 |
| AF347015.33 | -0.54 | -0.62 |
| BCL2L2 | -0.54 | -0.59 |
| GBP2 | -0.54 | -0.62 |
| HLA-F | -0.53 | -0.56 |
| AF347015.15 | -0.53 | -0.63 |
| MT-CYB | -0.53 | -0.61 |
| MT-CO2 | -0.53 | -0.59 |
| ARRDC2 | -0.52 | -0.66 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008415 | acyltransferase activity | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| AMUNDSON GENOTOXIC SIGNATURE | 105 | 68 | All SZGR 2.0 genes in this pathway |
| DAWSON METHYLATED IN LYMPHOMA TCL1 | 59 | 45 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-137 | 907 | 914 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-138 | 497 | 503 | 1A | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-182 | 927 | 933 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-188 | 452 | 458 | 1A | hsa-miR-188 | CAUCCCUUGCAUGGUGGAGGGU |
| miR-25/32/92/363/367 | 264 | 270 | m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-29 | 1015 | 1021 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-299-5p | 301 | 307 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU | ||||
| miR-329 | 371 | 377 | 1A | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
| hsa-miR-329brain | AACACACCUGGUUAACCUCUUU | ||||
| miR-369-3p | 399 | 405 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-370 | 499 | 506 | 1A,m8 | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-374 | 399 | 406 | 1A,m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-378 | 751 | 757 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| miR-485-5p | 1068 | 1074 | m8 | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
| miR-9 | 40 | 46 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-96 | 927 | 933 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.