Gene Page: APOBEC3C
Summary ?
| GeneID | 27350 |
| Symbol | APOBEC3C |
| Synonyms | A3C|APOBEC1L|ARDC2|ARDC4|ARP5|PBI|bK150C2.3 |
| Description | apolipoprotein B mRNA editing enzyme catalytic subunit 3C |
| Reference | MIM:607750|HGNC:HGNC:17353|HPRD:09671| |
| Gene type | protein-coding |
| Map location | 22q13.1 |
| Pascal p-value | 0.398 |
| Fetal beta | 0.056 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs138462 | chr22 | 38905445 | APOBEC3C | 27350 | 0.04 | cis | ||
| rs16999224 | chr22 | 39076586 | APOBEC3C | 27350 | 0.04 | cis | ||
| rs2716734 | chr2 | 39947720 | APOBEC3C | 27350 | 0.02 | trans | ||
| rs2716736 | chr2 | 39947946 | APOBEC3C | 27350 | 0.02 | trans | ||
| rs13024968 | chr2 | 60026328 | APOBEC3C | 27350 | 0.02 | trans | ||
| rs7430400 | chr3 | 65885662 | APOBEC3C | 27350 | 0 | trans | ||
| rs17023105 | chr4 | 96074164 | APOBEC3C | 27350 | 2.101E-4 | trans | ||
| rs10516959 | chr4 | 96088848 | APOBEC3C | 27350 | 0.06 | trans | ||
| rs7725925 | chr5 | 8275608 | APOBEC3C | 27350 | 0.2 | trans | ||
| rs996749 | chr5 | 8307934 | APOBEC3C | 27350 | 0.15 | trans | ||
| rs1587434 | chr6 | 66615354 | APOBEC3C | 27350 | 0.17 | trans | ||
| rs17165806 | chr7 | 93469599 | APOBEC3C | 27350 | 0.03 | trans | ||
| rs17165823 | chr7 | 93476735 | APOBEC3C | 27350 | 0.02 | trans | ||
| rs7777209 | chr7 | 93485280 | APOBEC3C | 27350 | 0.03 | trans | ||
| snp_a-2308001 | 0 | APOBEC3C | 27350 | 0.1 | trans | |||
| rs2294031 | chr8 | 1812077 | APOBEC3C | 27350 | 6.578E-4 | trans | ||
| rs870686 | chr8 | 25253521 | APOBEC3C | 27350 | 0.16 | trans | ||
| rs10503771 | chr8 | 25304019 | APOBEC3C | 27350 | 0.1 | trans | ||
| rs12352894 | chr9 | 38197383 | APOBEC3C | 27350 | 0.06 | trans | ||
| rs12255258 | chr10 | 106306927 | APOBEC3C | 27350 | 0.07 | trans | ||
| rs740597 | chr10 | 118508638 | APOBEC3C | 27350 | 0.1 | trans | ||
| rs11025109 | chr11 | 19405491 | APOBEC3C | 27350 | 0.11 | trans | ||
| rs7110934 | chr11 | 116127410 | APOBEC3C | 27350 | 0.01 | trans | ||
| rs7941495 | chr11 | 116129593 | APOBEC3C | 27350 | 0.04 | trans | ||
| rs7928297 | chr11 | 116129605 | APOBEC3C | 27350 | 0.04 | trans | ||
| rs17121555 | chr12 | 59241402 | APOBEC3C | 27350 | 0.01 | trans | ||
| rs17121606 | chr12 | 59258905 | APOBEC3C | 27350 | 0.01 | trans | ||
| rs1684198 | 0 | APOBEC3C | 27350 | 0.02 | trans | |||
| rs17075874 | chr13 | 22671340 | APOBEC3C | 27350 | 0.09 | trans | ||
| rs2764585 | chr13 | 46880234 | APOBEC3C | 27350 | 0.02 | trans | ||
| rs3742866 | 0 | APOBEC3C | 27350 | 0.18 | trans | |||
| rs4792093 | chr17 | 10939374 | APOBEC3C | 27350 | 0.07 | trans | ||
| rs7223509 | chr17 | 10940048 | APOBEC3C | 27350 | 0.07 | trans | ||
| rs3786474 | chr18 | 55135672 | APOBEC3C | 27350 | 0.17 | trans | ||
| rs1343409 | chrX | 104143804 | APOBEC3C | 27350 | 0.07 | trans | ||
| rs10521889 | chrX | 149836861 | APOBEC3C | 27350 | 0.03 | trans | ||
| rs1882710 | chrX | 149894355 | APOBEC3C | 27350 | 0.18 | trans |
Section II. Transcriptome annotation
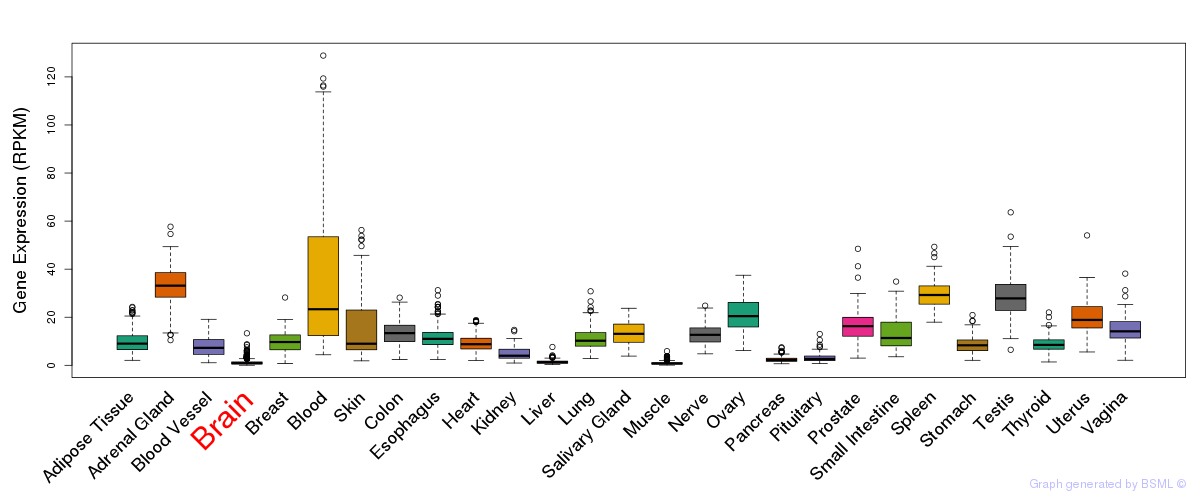
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BENNETT SYSTEMIC LUPUS ERYTHEMATOSUS | 31 | 21 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR DN | 86 | 62 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS UP | 63 | 36 | All SZGR 2.0 genes in this pathway |
| CROONQUIST IL6 DEPRIVATION DN | 98 | 69 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS SIGNALING DN | 72 | 47 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION DN | 99 | 65 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR DN | 37 | 20 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |