Gene Page: GNAS
Summary ?
| GeneID | 2778 |
| Symbol | GNAS |
| Synonyms | AHO|C20orf45|GNAS1|GPSA|GSA|GSP|NESP|POH|SCG6|SgVI |
| Description | GNAS complex locus |
| Reference | MIM:139320|HGNC:HGNC:4392|Ensembl:ENSG00000087460|HPRD:00761|Vega:OTTHUMG00000033069 |
| Gene type | protein-coding |
| Map location | 20q13.3 |
| Pascal p-value | 0.027 |
| Sherlock p-value | 0.626 |
| DEG p-value | DEG:Zhao_2015:p=6.55e-04:q=0.0955 |
| Fetal beta | -1.603 |
| DMG | 1 (# studies) |
| Support | DOPAMINE G-PROTEIN RELAY G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Zhao_2015 | RNA Sequencing analysis | Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0086 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GNAS | chr20 | 57466823 | C | T | NM_000516 NM_001077488 NM_001077489 NM_001077490 NM_016592 NM_080425 NM_080426 NR_003259 | . . . . . . . . | silent silent silent intronic intronic intronic silent intronic | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23143233 | 20 | 57465864 | GNAS | 2.87E-9 | -0.013 | 2.02E-6 | DMG:Jaffe_2016 |
| cg12372477 | 20 | 57465915 | GNAS | 1.64E-8 | -0.008 | 6.03E-6 | DMG:Jaffe_2016 |
| cg21330323 | 20 | 57414596 | GNAS | 9.29E-8 | -0.015 | 2.11E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
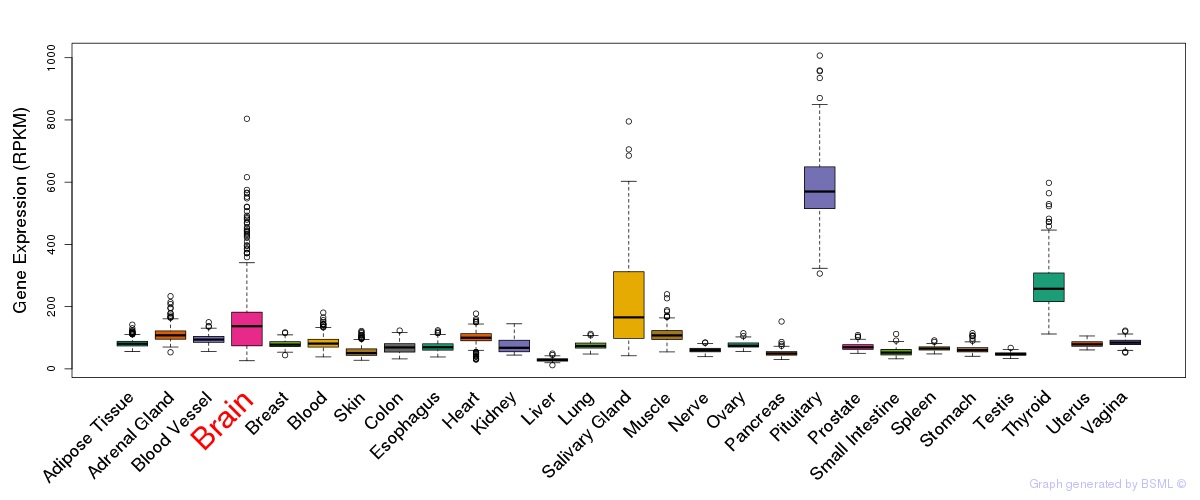
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004871 | signal transducer activity | IEA | - | |
| GO:0003674 | molecular_function | ND | - | |
| GO:0003924 | GTPase activity | NAS | 1716359 | |
| GO:0003924 | GTPase activity | TAS | 9159128 | |
| GO:0005525 | GTP binding | IEA | - | |
| GO:0005525 | GTP binding | NAS | - | |
| GO:0019001 | guanyl nucleotide binding | IEA | - | |
| GO:0042802 | identical protein binding | IPI | 17353931 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007191 | dopamine receptor, adenylate cyclase activating pathway | IEA | dopamine (GO term level: 12) | - |
| GO:0001958 | endochondral ossification | IEA | - | |
| GO:0007190 | activation of adenylate cyclase activity | TAS | 9159128 | |
| GO:0007189 | G-protein signaling, adenylate cyclase activating pathway | NAS | 1716359 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0006112 | energy reserve metabolic process | IEA | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0007165 | signal transduction | NAS | - | |
| GO:0009306 | protein secretion | NAS | 10729789 | |
| GO:0009755 | hormone-mediated signaling | EXP | 12626323 | |
| GO:0009791 | post-embryonic development | IEA | - | |
| GO:0007608 | sensory perception of smell | TAS | 3018580 | |
| GO:0007565 | female pregnancy | NAS | 10729789 | |
| GO:0006605 | protein targeting | IEA | - | |
| GO:0042493 | response to drug | IEA | - | |
| GO:0051216 | cartilage development | IEA | - | |
| GO:0040032 | post-embryonic body morphogenesis | IEA | - | |
| GO:0048701 | embryonic cranial skeleton morphogenesis | IEA | - | |
| GO:0035116 | embryonic hindlimb morphogenesis | IEA | - | |
| GO:0046907 | intracellular transport | NAS | 7997272 | |
| GO:0045669 | positive regulation of osteoblast differentiation | IEA | - | |
| GO:0045672 | positive regulation of osteoclast differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042995 | cell projection | IEA | axon (GO term level: 4) | - |
| GO:0005834 | heterotrimeric G-protein complex | NAS | 1716359 | |
| GO:0005834 | heterotrimeric G-protein complex | TAS | 9159128 | |
| GO:0005576 | extracellular region | NAS | - | |
| GO:0005624 | membrane fraction | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 11256614 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005886 | plasma membrane | NAS | - | |
| GO:0005886 | plasma membrane | TAS | 9727013 | |
| GO:0031224 | intrinsic to membrane | IDA | 7997272 | |
| GO:0032588 | trans-Golgi network membrane | IDA | 7997272 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADCY2 | AC2 | FLJ16822 | FLJ45092 | HBAC2 | KIAA1060 | MGC133314 | adenylate cyclase 2 (brain) | - | HPRD | 9268375 |
| ADCY5 | AC5 | adenylate cyclase 5 | - | HPRD | 9268375 |
| ADCY6 | AC6 | DKFZp779F075 | KIAA0422 | adenylate cyclase 6 | - | HPRD,BioGRID | 9228084 |
| ADORA1 | RDC7 | adenosine A1 receptor | - | HPRD | 10521440 |
| ADRB3 | BETA3AR | adrenergic, beta-3-, receptor | - | HPRD | 8011597 |
| AVPR2 | ADHR | DI1 | DIR | DIR3 | MGC126533 | MGC138386 | NDI | V2R | arginine vasopressin receptor 2 | - | HPRD | 8621513 |
| CAV3 | LGMD1C | LQT9 | MGC126100 | MGC126101 | MGC126129 | VIP-21 | VIP21 | caveolin 3 | - | HPRD,BioGRID | 7797570 |
| CRHR1 | CRF-R | CRF1 | CRFR1 | CRH-R1h | CRHR | CRHR1f | corticotropin releasing hormone receptor 1 | - | HPRD,BioGRID | 10598591 |
| FSCN1 | FLJ38511 | SNL | p55 | fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) | Affinity Capture-MS | BioGRID | 17353931 |
| GCGR | GGR | MGC138246 | glucagon receptor | - | HPRD | 1908089 |
| GNB1 | - | guanine nucleotide binding protein (G protein), beta polypeptide 1 | G-beta-1 interacts with G-alpha-s. | BIND | 15782186 |
| GNB1 | - | guanine nucleotide binding protein (G protein), beta polypeptide 1 | - | HPRD | 11294873 |
| HTR6 | 5-HT6 | 5-HT6R | 5-hydroxytryptamine (serotonin) receptor 6 | - | HPRD | 11916537 |
| LHCGR | FLJ41504 | LCGR | LGR2 | LH/CG-R | LH/CGR | LHR | LHRHR | LSH-R | luteinizing hormone/choriogonadotropin receptor | - | HPRD | 10493819 |
| PAICS | ADE2 | ADE2H1 | AIRC | DKFZp781N1372 | MGC1343 | MGC5024 | PAIS | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | Affinity Capture-MS | BioGRID | 17353931 |
| PLEKHB1 | KPL1 | PHR1 | PHRET1 | pleckstrin homology domain containing, family B (evectins) member 1 | - | HPRD | 10585447 |
| PTGDR | AS1 | ASRT1 | DP | DP1 | MGC49004 | prostaglandin D2 receptor (DP) | - | HPRD,BioGRID | 12672054 |
| PTGIR | IP | MGC102830 | PRIPR | prostaglandin I2 (prostacyclin) receptor (IP) | - | HPRD,BioGRID | 12488443 |
| RGS2 | G0S8 | regulator of G-protein signaling 2, 24kDa | - | HPRD,BioGRID | 9794454 |
| RIC8A | MGC104517 | MGC131931 | MGC148073 | MGC148074 | RIC8 | synembryn | resistance to inhibitors of cholinesterase 8 homolog A (C. elegans) | - | HPRD,BioGRID | 12652642 |
| RIC8B | FLJ10620 | MGC39476 | RIC8 | hSyn | resistance to inhibitors of cholinesterase 8 homolog B (C. elegans) | - | HPRD | 12652642 |
| RUVBL1 | ECP54 | INO80H | NMP238 | PONTIN | Pontin52 | RVB1 | TIH1 | TIP49 | TIP49A | RuvB-like 1 (E. coli) | Affinity Capture-MS | BioGRID | 17353931 |
| SHMT2 | GLYA | SHMT | serine hydroxymethyltransferase 2 (mitochondrial) | Affinity Capture-MS | BioGRID | 17353931 |
| SNX13 | KIAA0713 | RGS-PX1 | sorting nexin 13 | - | HPRD,BioGRID | 11729322 |
| TSHR | CHNG1 | LGR3 | MGC75129 | hTSHR-I | thyroid stimulating hormone receptor | - | HPRD | 9525885 |
| TTC1 | FLJ46404 | TPR1 | tetratricopeptide repeat domain 1 | G-alpha-s-Short interacts with TPR1. | BIND | 12748287 |
| TTC1 | FLJ46404 | TPR1 | tetratricopeptide repeat domain 1 | - | HPRD,BioGRID | 12748287 |
| TTC1 | FLJ46404 | TPR1 | tetratricopeptide repeat domain 1 | G-alpha-s-Long interacts with TPR1. | BIND | 12748287 |
| VIPR1 | FLJ41949 | HVR1 | II | PACAP-R-2 | RDC1 | VAPC1 | VIPR | VIRG | VPAC1 | VPCAP1R | vasoactive intestinal peptide receptor 1 | - | HPRD,BioGRID | 11812005 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG GAP JUNCTION | 90 | 68 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG TASTE TRANSDUCTION | 52 | 28 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOGENESIS | 102 | 80 | All SZGR 2.0 genes in this pathway |
| KEGG VASOPRESSIN REGULATED WATER REABSORPTION | 44 | 30 | All SZGR 2.0 genes in this pathway |
| KEGG VIBRIO CHOLERAE INFECTION | 56 | 32 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CSK PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AGPCR PATHWAY | 13 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CCR3 PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GCR PATHWAY | 22 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CFTR PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERK PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MPR PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PLCE PATHWAY | 12 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BARR MAPK PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BARRESTIN SRC PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GPCR PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BARRESTIN PATHWAY | 10 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CREB PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | 34 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | 43 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | 88 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCAGON TYPE LIGAND RECEPTORS | 33 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA S SIGNALLING EVENTS | 121 | 82 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Z SIGNALLING EVENTS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET HOMEOSTASIS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME AQUAPORIN MEDIATED TRANSPORT | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | 19 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| COLDREN GEFITINIB RESISTANCE DN | 230 | 115 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP | 117 | 85 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN | 137 | 97 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| TOMIDA METASTASIS UP | 26 | 13 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION UP | 60 | 45 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS UP | 110 | 71 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| JOHANSSON BRAIN CANCER EARLY VS LATE DN | 45 | 35 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| ALCALA APOPTOSIS | 88 | 60 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 8 | 36 | 28 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 20Q12 Q13 AMPLICON | 149 | 76 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED SIGNIFICANTLY | 26 | 22 | All SZGR 2.0 genes in this pathway |
| GNATENKO PLATELET SIGNATURE | 48 | 28 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA PROGNOSIS UP | 47 | 30 | All SZGR 2.0 genes in this pathway |
| LU IL4 SIGNALING | 94 | 56 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION UP | 39 | 29 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA UP | 98 | 64 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS RESPONSE | 35 | 28 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD DN | 150 | 93 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY DN | 145 | 88 | All SZGR 2.0 genes in this pathway |
| ZHANG PROLIFERATING VS QUIESCENT | 51 | 41 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN | 102 | 65 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| VANASSE BCL2 TARGETS UP | 40 | 25 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER WITH EPCAM UP | 53 | 25 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| NIELSEN MALIGNAT FIBROUS HISTIOCYTOMA DN | 18 | 11 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| RAGHAVACHARI PLATELET SPECIFIC GENES | 70 | 46 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY TARGETS OF MUTATED TP53 UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS UP | 139 | 93 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| BRIDEAU IMPRINTED GENES | 63 | 47 | All SZGR 2.0 genes in this pathway |