Gene Page: GPR1
Summary ?
| GeneID | 2825 |
| Symbol | GPR1 |
| Synonyms | - |
| Description | G protein-coupled receptor 1 |
| Reference | MIM:600239|HGNC:HGNC:4463|Ensembl:ENSG00000183671|HPRD:02585|Vega:OTTHUMG00000132894 |
| Gene type | protein-coding |
| Map location | 2q33.3 |
| Pascal p-value | 0.01 |
| Sherlock p-value | 0.658 |
| Fetal beta | -0.005 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00916 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01016 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
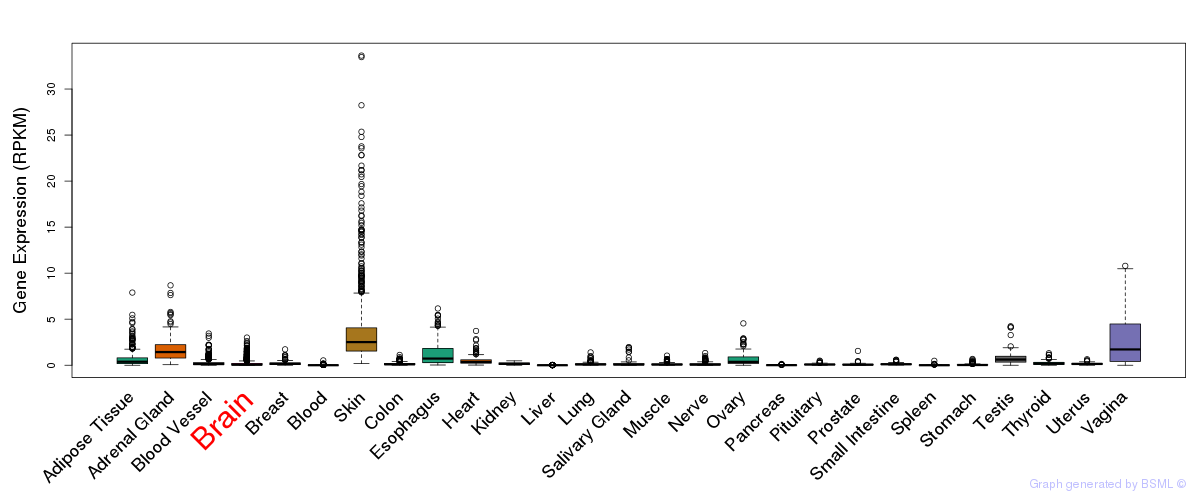
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RPP30 | 0.77 | 0.83 |
| PSMD13 | 0.77 | 0.82 |
| PSMB7 | 0.76 | 0.80 |
| OCIAD2 | 0.75 | 0.85 |
| CCT7 | 0.75 | 0.80 |
| GLA | 0.75 | 0.76 |
| EIF2B4 | 0.75 | 0.81 |
| EIF6 | 0.75 | 0.79 |
| IP6K2 | 0.75 | 0.82 |
| UFD1L | 0.75 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.33 | -0.64 | -0.70 |
| AF347015.27 | -0.63 | -0.70 |
| AF347015.8 | -0.63 | -0.69 |
| TINAGL1 | -0.62 | -0.69 |
| AF347015.15 | -0.62 | -0.70 |
| SLC6A12 | -0.62 | -0.72 |
| HLA-F | -0.62 | -0.68 |
| AF347015.2 | -0.62 | -0.70 |
| MT-CO2 | -0.61 | -0.66 |
| APOL1 | -0.61 | -0.72 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | TAS | 7851889 | |
| GO:0007165 | signal transduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 7851889 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| SU PANCREAS | 54 | 30 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| LEIN MIDBRAIN MARKERS | 82 | 55 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| WINZEN DEGRADED VIA KHSRP | 100 | 70 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |