Gene Page: GRIA2
Summary ?
| GeneID | 2891 |
| Symbol | GRIA2 |
| Synonyms | GLUR2|GLURB|GluA2|GluR-K2|HBGR2 |
| Description | glutamate ionotropic receptor AMPA type subunit 2 |
| Reference | MIM:138247|HGNC:HGNC:4572|Ensembl:ENSG00000120251|HPRD:04726|Vega:OTTHUMG00000133836 |
| Gene type | protein-coding |
| Map location | 4q32.1 |
| Pascal p-value | 0.235 |
| Sherlock p-value | 0.755 |
| Fetal beta | -0.172 |
| Support | LIGAND GATED ION SIGNALING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanARC G2Cdb.humanPSD G2Cdb.humanPSP Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 6 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.6626 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
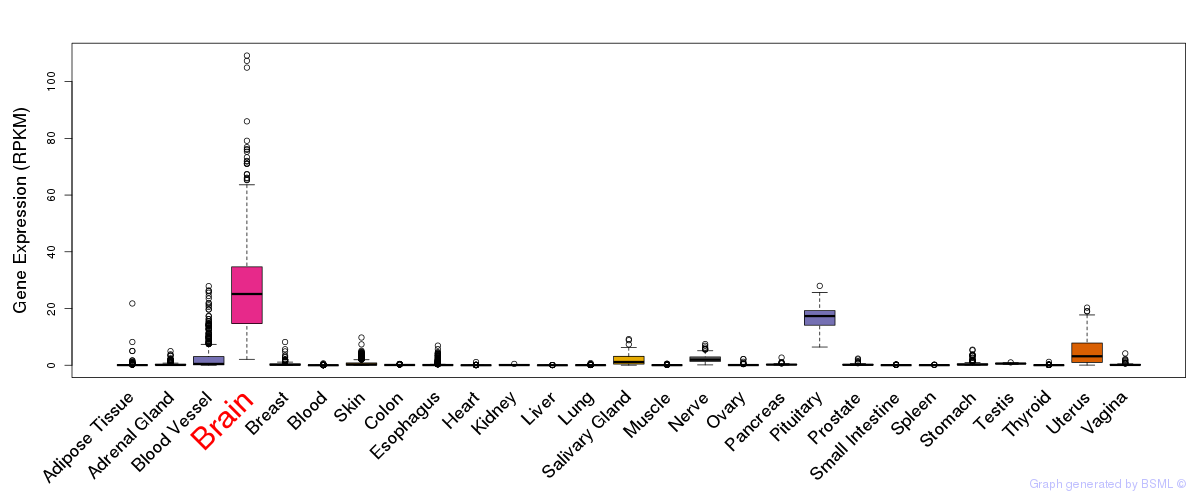
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TACC1 | 0.94 | 0.89 |
| ZMAT3 | 0.93 | 0.88 |
| FAM73A | 0.92 | 0.81 |
| CHIC1 | 0.91 | 0.80 |
| MBNL2 | 0.90 | 0.87 |
| EFR3A | 0.90 | 0.79 |
| DPP8 | 0.90 | 0.78 |
| NAPEPLD | 0.90 | 0.77 |
| OXR1 | 0.89 | 0.79 |
| ARAP2 | 0.89 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.48 | -0.65 |
| TBC1D10A | -0.48 | -0.44 |
| TRAF4 | -0.46 | -0.63 |
| AC006276.2 | -0.46 | -0.52 |
| SH3BP2 | -0.46 | -0.57 |
| SH2B2 | -0.45 | -0.56 |
| RPL36 | -0.45 | -0.65 |
| RPLP1 | -0.45 | -0.61 |
| SIGIRR | -0.45 | -0.57 |
| NR2C2AP | -0.45 | -0.50 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004971 | alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity | TAS | glutamate (GO term level: 8) | 8003671 |
| GO:0004970 | ionotropic glutamate receptor activity | IEA | glutamate (GO term level: 7) | - |
| GO:0005234 | extracellular-glutamate-gated ion channel activity | IEA | glutamate (GO term level: 11) | - |
| GO:0015277 | kainate selective glutamate receptor activity | TAS | glutamate (GO term level: 8) | 8003671 |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005216 | ion channel activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 8003671 |
| GO:0007165 | signal transduction | TAS | 8003671 | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0006810 | transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | TAS | 8003671 | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CACNG2 | MGC138502 | MGC138504 | calcium channel, voltage-dependent, gamma subunit 2 | Affinity Capture-Western | BioGRID | 11140673 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | Affinity Capture-MS | BioGRID | 16990550 |
| GRIA1 | GLUH1 | GLUR1 | GLURA | HBGR1 | MGC133252 | glutamate receptor, ionotropic, AMPA 1 | Affinity Capture-Western | BioGRID | 10358037 |
| GRID2 | MGC117022 | MGC117023 | MGC117024 | glutamate receptor, ionotropic, delta 2 | Affinity Capture-Western | BioGRID | 12573530 |
| GRIK2 | EAA4 | GLR6 | GLUK6 | GLUR6 | MGC74427 | MRT6 | glutamate receptor, ionotropic, kainate 2 | Affinity Capture-Western | BioGRID | 8288598 |
| GRIK5 | EAA2 | GRIK2 | KA2 | glutamate receptor, ionotropic, kainate 5 | Affinity Capture-Western | BioGRID | 12573530 |
| GRIP1 | GRIP | glutamate receptor interacting protein 1 | Reconstituted Complex Two-hybrid | BioGRID | 11891216 |
| PICK1 | MGC15204 | PICK | PRKCABP | protein interacting with PRKCA 1 | Reconstituted Complex Two-hybrid | BioGRID | 11891216 |
| SDCBP | MDA-9 | ST1 | SYCL | TACIP18 | syndecan binding protein (syntenin) | Reconstituted Complex | BioGRID | 11891216 |
| SPTAN1 | (ALPHA)II-SPECTRIN | FLJ44613 | NEAS | spectrin, alpha, non-erythrocytic 1 (alpha-fodrin) | Affinity Capture-Western Far Western | BioGRID | 10576550 |
| SPTBN1 | ELF | SPTB2 | betaSpII | spectrin, beta, non-erythrocytic 1 | Far Western | BioGRID | 10576550 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| PID NCADHERIN PATHWAY | 33 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAFFICKING OF AMPA RECEPTORS | 28 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | 16 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | 15 | 14 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL UP | 94 | 59 | All SZGR 2.0 genes in this pathway |
| GUENTHER GROWTH SPHERICAL VS ADHERENT UP | 21 | 15 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN HIGHEST EXPRESSION | 40 | 29 | All SZGR 2.0 genes in this pathway |
| ABBUD LIF SIGNALING 1 DN | 26 | 17 | All SZGR 2.0 genes in this pathway |
| MCCLUNG DELTA FOSB TARGETS 8WK | 47 | 38 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS POSTNATAL | 63 | 50 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX UP | 36 | 27 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS DN | 74 | 55 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| LEIN NEURON MARKERS | 69 | 45 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS DN | 120 | 81 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 188 | 194 | m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 187 | 194 | 1A,m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-132/212 | 2343 | 2349 | 1A | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-181 | 140 | 146 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-186 | 779 | 786 | 1A,m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-218 | 476 | 482 | m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-219 | 1988 | 1994 | 1A | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-30-5p | 2417 | 2424 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-335 | 456 | 462 | 1A | hsa-miR-335brain | UCAAGAGCAAUAACGAAAAAUGU |
| miR-374 | 1284 | 1290 | 1A | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-410 | 2097 | 2103 | m8 | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-486 | 48 | 54 | 1A | hsa-miR-486 | UCCUGUACUGAGCUGCCCCGAG |
| miR-496 | 2090 | 2096 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| hsa-miR-496 | AUUACAUGGCCAAUCUC | ||||
| hsa-miR-496 | AUUACAUGGCCAAUCUC | ||||
| hsa-miR-496 | AUUACAUGGCCAAUCUC | ||||
| hsa-miR-496 | AUUACAUGGCCAAUCUC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.