Gene Page: SLC2A8
Summary ?
| GeneID | 29988 |
| Symbol | SLC2A8 |
| Synonyms | GLUT8|GLUTX1 |
| Description | solute carrier family 2 member 8 |
| Reference | MIM:605245|HGNC:HGNC:13812|Ensembl:ENSG00000136856|HPRD:05580|Vega:OTTHUMG00000020702 |
| Gene type | protein-coding |
| Map location | 9q33.3 |
| Pascal p-value | 0.006 |
| Sherlock p-value | 0.875 |
| Fetal beta | -0.581 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03804985 | 9 | 130159694 | SLC2A8 | -0.027 | 0.91 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7044100 | chr9 | 129742639 | SLC2A8 | 29988 | 0.05 | cis | ||
| rs12644099 | 0 | SLC2A8 | 29988 | 0.11 | trans | |||
| rs1138739 | 9 | 130164993 | SLC2A8 | ENSG00000136856.13 | 2.389E-6 | 0.02 | 5572 | gtex_brain_ba24 |
| rs1138740 | 9 | 130165995 | SLC2A8 | ENSG00000136856.13 | 1.55E-6 | 0.02 | 6574 | gtex_brain_ba24 |
| rs11788710 | 9 | 130166383 | SLC2A8 | ENSG00000136856.13 | 1.99E-6 | 0.02 | 6962 | gtex_brain_ba24 |
| rs11790936 | 9 | 130170526 | SLC2A8 | ENSG00000136856.13 | 4.052E-6 | 0.02 | 11105 | gtex_brain_ba24 |
| rs11790967 | 9 | 130170561 | SLC2A8 | ENSG00000136856.13 | 2.009E-6 | 0.02 | 11140 | gtex_brain_ba24 |
| rs11794990 | 9 | 130171020 | SLC2A8 | ENSG00000136856.13 | 2.02E-6 | 0.02 | 11599 | gtex_brain_ba24 |
| rs10987639 | 9 | 130172485 | SLC2A8 | ENSG00000136856.13 | 1.985E-6 | 0.02 | 13064 | gtex_brain_ba24 |
| rs11791288 | 9 | 130173224 | SLC2A8 | ENSG00000136856.13 | 2.009E-6 | 0.02 | 13803 | gtex_brain_ba24 |
| rs11791717 | 9 | 130173320 | SLC2A8 | ENSG00000136856.13 | 2.016E-6 | 0.02 | 13899 | gtex_brain_ba24 |
| rs7873847 | 9 | 130174444 | SLC2A8 | ENSG00000136856.13 | 2.18E-6 | 0.02 | 15023 | gtex_brain_ba24 |
| rs10987643 | 9 | 130175263 | SLC2A8 | ENSG00000136856.13 | 2.475E-6 | 0.02 | 15842 | gtex_brain_ba24 |
| rs4837147 | 9 | 130125645 | SLC2A8 | ENSG00000136856.13 | 1.169E-6 | 0 | -33776 | gtex_brain_putamen_basal |
| rs10987619 | 9 | 130131996 | SLC2A8 | ENSG00000136856.13 | 1.226E-6 | 0 | -27425 | gtex_brain_putamen_basal |
| rs1138739 | 9 | 130164993 | SLC2A8 | ENSG00000136856.13 | 1.961E-7 | 0 | 5572 | gtex_brain_putamen_basal |
| rs1138740 | 9 | 130165995 | SLC2A8 | ENSG00000136856.13 | 1.496E-7 | 0 | 6574 | gtex_brain_putamen_basal |
| rs11788710 | 9 | 130166383 | SLC2A8 | ENSG00000136856.13 | 7.95E-8 | 0 | 6962 | gtex_brain_putamen_basal |
| rs11790936 | 9 | 130170526 | SLC2A8 | ENSG00000136856.13 | 5.43E-8 | 0 | 11105 | gtex_brain_putamen_basal |
| rs11790967 | 9 | 130170561 | SLC2A8 | ENSG00000136856.13 | 2.845E-8 | 0 | 11140 | gtex_brain_putamen_basal |
| rs11794990 | 9 | 130171020 | SLC2A8 | ENSG00000136856.13 | 2.845E-8 | 0 | 11599 | gtex_brain_putamen_basal |
| rs151232558 | 9 | 130171848 | SLC2A8 | ENSG00000136856.13 | 2.301E-8 | 0 | 12427 | gtex_brain_putamen_basal |
| rs28546643 | 9 | 130171859 | SLC2A8 | ENSG00000136856.13 | 4.992E-7 | 0 | 12438 | gtex_brain_putamen_basal |
| rs10987639 | 9 | 130172485 | SLC2A8 | ENSG00000136856.13 | 2.845E-8 | 0 | 13064 | gtex_brain_putamen_basal |
| rs11791288 | 9 | 130173224 | SLC2A8 | ENSG00000136856.13 | 2.838E-8 | 0 | 13803 | gtex_brain_putamen_basal |
| rs11791717 | 9 | 130173320 | SLC2A8 | ENSG00000136856.13 | 2.835E-8 | 0 | 13899 | gtex_brain_putamen_basal |
| rs7873847 | 9 | 130174444 | SLC2A8 | ENSG00000136856.13 | 2.81E-8 | 0 | 15023 | gtex_brain_putamen_basal |
| rs10987643 | 9 | 130175263 | SLC2A8 | ENSG00000136856.13 | 2.782E-8 | 0 | 15842 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
General gene expression (GTEx)
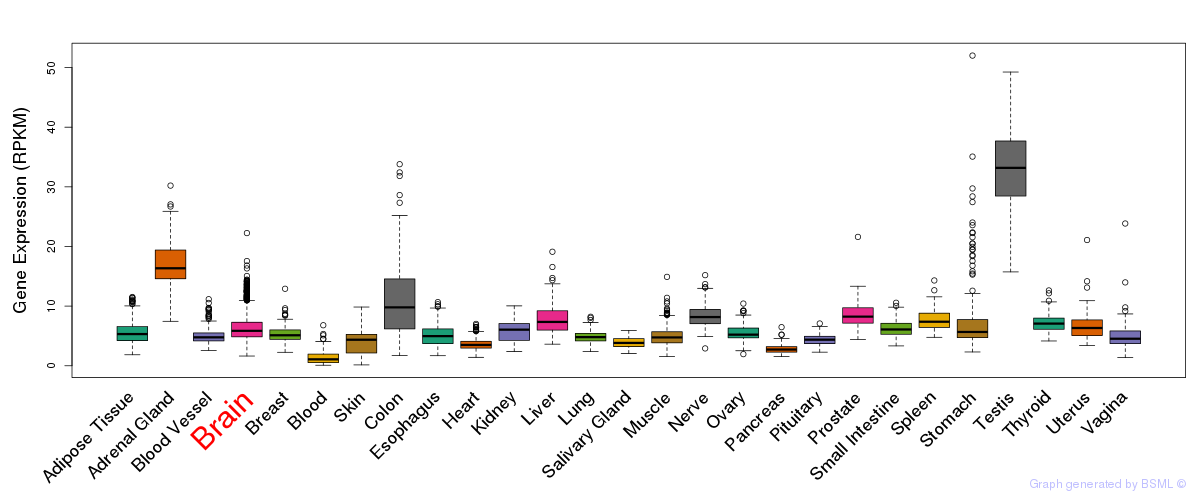
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT | 241 | 157 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | 89 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | 12 | 10 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER WITH LOH IN CHR9Q | 116 | 71 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS DN | 120 | 69 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P2 | 79 | 55 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE DN | 138 | 99 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |