Gene Page: APEX1
Summary ?
| GeneID | 328 |
| Symbol | APEX1 |
| Synonyms | APE|APE1|APEN|APEX|APX|HAP1|REF1 |
| Description | apurinic/apyrimidinic endodeoxyribonuclease 1 |
| Reference | MIM:107748|HGNC:HGNC:587|Ensembl:ENSG00000100823|HPRD:00136|Vega:OTTHUMG00000029544 |
| Gene type | protein-coding |
| Map location | 14q11.2 |
| Pascal p-value | 0.716 |
| Sherlock p-value | 0.724 |
| Fetal beta | 1.364 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.047 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0385 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22438324 | 14 | 20922915 | APEX1 | -0.023 | 0.34 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
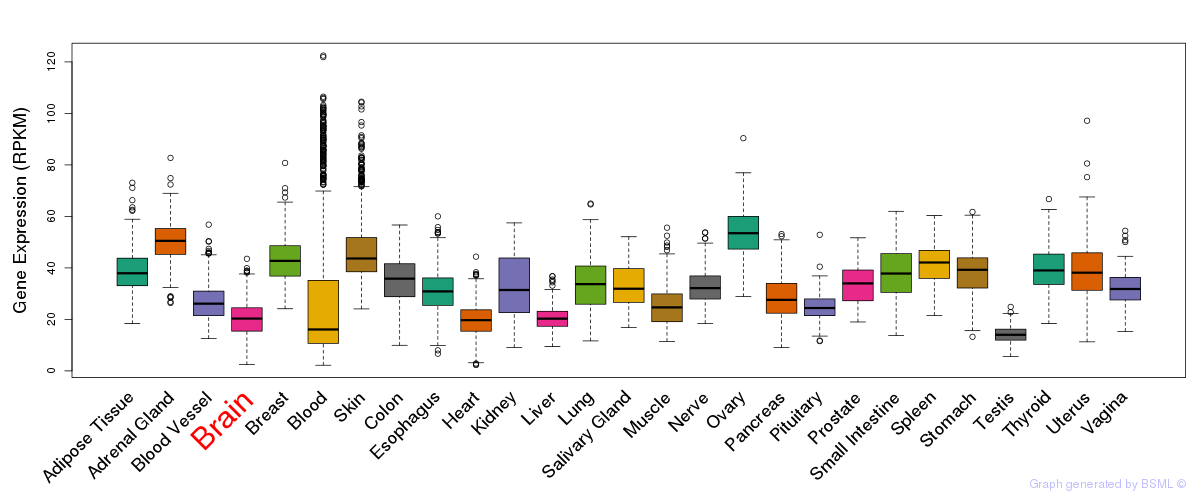
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HOOK3 | 0.92 | 0.92 |
| UTP14C | 0.91 | 0.92 |
| TMF1 | 0.91 | 0.93 |
| ZNF791 | 0.89 | 0.91 |
| CLPX | 0.89 | 0.90 |
| RABGAP1 | 0.89 | 0.91 |
| ATRX | 0.89 | 0.91 |
| C9orf102 | 0.89 | 0.91 |
| SEC63 | 0.89 | 0.90 |
| UGCGL1 | 0.88 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.68 | -0.73 |
| FXYD1 | -0.67 | -0.72 |
| HIGD1B | -0.67 | -0.73 |
| MT-CO2 | -0.67 | -0.72 |
| ENHO | -0.66 | -0.77 |
| AF347015.21 | -0.66 | -0.72 |
| IFI27 | -0.66 | -0.72 |
| METRN | -0.64 | -0.71 |
| CST3 | -0.63 | -0.70 |
| AF347015.27 | -0.63 | -0.67 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0003677 | DNA binding | IDA | 11286553 | |
| GO:0003713 | transcription coactivator activity | IDA | 9119221 | |
| GO:0003714 | transcription corepressor activity | TAS | 7961715 | |
| GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity | TAS | 9119221 | |
| GO:0005515 | protein binding | IPI | 12524539 |15518571 | |
| GO:0004844 | uracil DNA N-glycosylase activity | TAS | 10805771 | |
| GO:0004520 | endodeoxyribonuclease activity | TAS | 1722334 | |
| GO:0004523 | ribonuclease H activity | TAS | 11286553 | |
| GO:0004528 | phosphodiesterase I activity | TAS | 9119221 | |
| GO:0016829 | lyase activity | IEA | - | |
| GO:0008408 | 3'-5' exonuclease activity | TAS | 11286553 | |
| GO:0016491 | oxidoreductase activity | IDA | 9119221 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006366 | transcription from RNA polymerase II promoter | TAS | 1380454 | |
| GO:0006284 | base-excision repair | TAS | 11286553 | |
| GO:0051101 | regulation of DNA binding | IDA | 9119221 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005813 | centrosome | IDA | 18029348 | |
| GO:0005840 | ribosome | TAS | 12524539 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IDA | 9119221 | |
| GO:0005654 | nucleoplasm | EXP | 9207062 |10559261 |11250913 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005783 | endoplasmic reticulum | TAS | 12524539 | |
| GO:0048471 | perinuclear region of cytoplasm | IDA | 12524539 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANP32A | C15orf1 | I1PP2A | LANP | MAPM | MGC119787 | MGC150373 | PHAP1 | PHAPI | PP32 | acidic (leucine-rich) nuclear phosphoprotein 32 family, member A | Affinity Capture-Western | BioGRID | 12524539 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | Ref1 interacts weakly with p21 promoter. | BIND | 15674341 |
| DCTN1 | DAP-150 | DP-150 | HMN7B | P135 | dynactin 1 (p150, glued homolog, Drosophila) | - | HPRD | 9361024 |
| FEN1 | FEN-1 | MF1 | RAD2 | flap structure-specific endonuclease 1 | - | HPRD,BioGRID | 11601988 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | - | HPRD,BioGRID | 10594042 |
| HMGB2 | HMG2 | high-mobility group box 2 | - | HPRD | 12524539 |
| HNRNPL | FLJ35509 | HNRPL | P/OKcl.14 | hnRNP-L | heterogeneous nuclear ribonucleoprotein L | - | HPRD,BioGRID | 11809897 |
| HSPA1A | FLJ54303 | FLJ54370 | FLJ54392 | FLJ54408 | FLJ75127 | HSP70-1 | HSP70-1A | HSP70I | HSP72 | HSPA1 | HSPA1B | heat shock 70kDa protein 1A | - | HPRD | 11133992 |
| MUTYH | MGC4416 | MYH | mutY homolog (E. coli) | hMYH is associated in vivo with apurinic/apyrimidinic endonuclease (APE1) | BIND | 11092888 |
| MUTYH | MGC4416 | MYH | mutY homolog (E. coli) | - | HPRD,BioGRID | 11092888 |
| NME1 | AWD | GAAD | NB | NBS | NDPK-A | NDPKA | NM23 | NM23-H1 | non-metastatic cells 1, protein (NM23A) expressed in | - | HPRD | 12524539 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | - | HPRD,BioGRID | 11601988 |
| SET | 2PP2A | I2PP2A | IGAAD | IPP2A2 | PHAPII | TAF-I | TAF-IBETA | SET nuclear oncogene | - | HPRD | 12628186 |
| SET | 2PP2A | I2PP2A | IGAAD | IPP2A2 | PHAPII | TAF-I | TAF-IBETA | SET nuclear oncogene | Affinity Capture-Western | BioGRID | 12524539 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with Ref-1. | BIND | 15824742 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 9119221 |
| TXN | DKFZp686B1993 | MGC61975 | TRX | TRX1 | thioredoxin | - | HPRD | 10585464|11118054 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | - | HPRD,BioGRID | 12110916 |
| XRCC1 | RCC | X-ray repair complementing defective repair in Chinese hamster cells 1 | - | HPRD,BioGRID | 11707423 |
| XRCC5 | FLJ39089 | KARP-1 | KARP1 | KU80 | KUB2 | Ku86 | NFIV | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) | Reconstituted Complex | BioGRID | 8621488 |
| XRCC6 | CTC75 | CTCBF | G22P1 | KU70 | ML8 | TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 | - | HPRD | 8621488 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG BASE EXCISION REPAIR | 35 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SET PATHWAY | 11 | 6 | All SZGR 2.0 genes in this pathway |
| PID HIF2PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME BASE EXCISION REPAIR | 19 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | 17 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | 10 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | 12 | 7 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| WAKASUGI HAVE ZNF143 BINDING SITES | 58 | 33 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| MENSSEN MYC TARGETS | 53 | 35 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS DN | 87 | 69 | All SZGR 2.0 genes in this pathway |
| SANA RESPONSE TO IFNG DN | 85 | 56 | All SZGR 2.0 genes in this pathway |
| FERRANDO LYL1 NEIGHBORS | 15 | 12 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT DN | 129 | 86 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS DN | 86 | 67 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 DN | 43 | 31 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER HEREDITARY VS SPORADIC | 50 | 32 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| HARRIS HYPOXIA | 81 | 64 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIN ACTION DIRECT VS INDIRECT 24HR | 55 | 38 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS INTERMEDIATE PROGENITOR | 149 | 84 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR DN | 101 | 64 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| IRITANI MAD1 TARGETS DN | 47 | 30 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| DELPUECH FOXO3 TARGETS DN | 41 | 27 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |