Gene Page: HSPG2
Summary ?
| GeneID | 3339 |
| Symbol | HSPG2 |
| Synonyms | HSPG|PLC|PRCAN|SJA|SJS|SJS1 |
| Description | heparan sulfate proteoglycan 2 |
| Reference | MIM:142461|HGNC:HGNC:5273|Ensembl:ENSG00000142798|HPRD:00804|Vega:OTTHUMG00000002674 |
| Gene type | protein-coding |
| Map location | 1p36.1-p34 |
| Pascal p-value | 0.447 |
| Fetal beta | 0.203 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Ambalavanan_2016 | Whole Exome Sequencing | This dataset includes 20 de novo mutations detected in 17 COS probands. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HSPG2 | chr1 | 22192291 | G | A | NM_005529 | p.(=) | synonymous SNV | Childhood-onset schizophrenia | DNM:Ambalavanan_2016 |
Section II. Transcriptome annotation
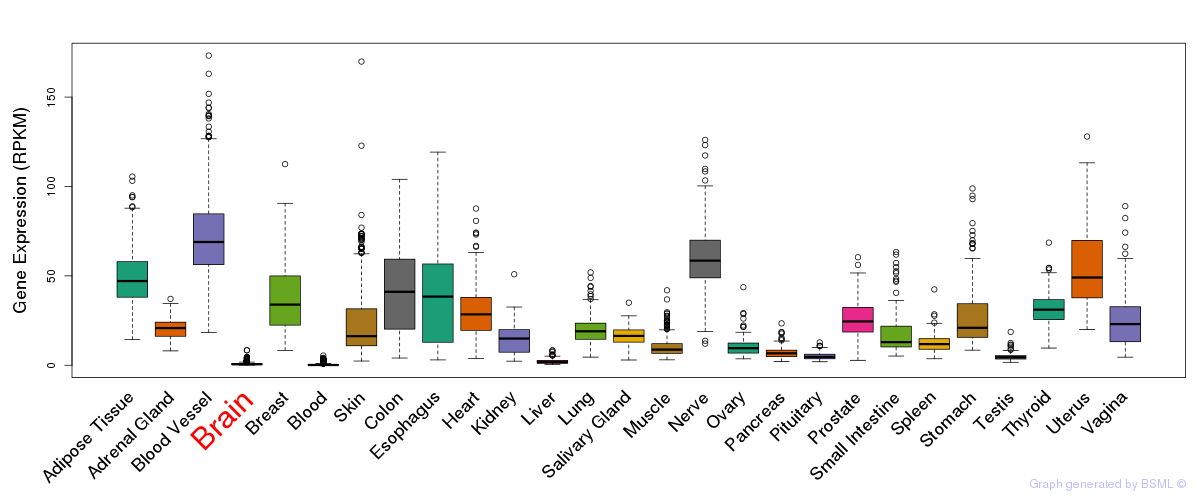
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KCNIP2 | 0.65 | 0.55 |
| HPCA | 0.60 | 0.53 |
| TNFAIP8L3 | 0.59 | 0.39 |
| SYTL5 | 0.58 | 0.55 |
| RIN1 | 0.56 | 0.46 |
| C1QTNF3 | 0.56 | 0.51 |
| TOMM34 | 0.56 | 0.50 |
| F12 | 0.56 | 0.62 |
| PNCK | 0.55 | 0.59 |
| C20orf103 | 0.55 | 0.38 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.23 | -0.29 |
| AF347015.18 | -0.23 | -0.30 |
| AF347015.2 | -0.23 | -0.27 |
| MT-ATP8 | -0.22 | -0.28 |
| ANKRD18B | -0.21 | -0.21 |
| MT-CO2 | -0.19 | -0.28 |
| AF347015.8 | -0.19 | -0.26 |
| TES | -0.18 | -0.09 |
| NOSTRIN | -0.18 | -0.25 |
| AF347015.15 | -0.18 | -0.25 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | - | HPRD | 9136074 |
| COL13A1 | COLXIIIA1 | FLJ42485 | collagen, type XIII, alpha 1 | - | HPRD,BioGRID | 11956183 |
| FBLN1 | FBLN | fibulin 1 | - | HPRD,BioGRID | 7500359 |
| FBLN2 | - | fibulin 2 | - | HPRD | 9431988 |
| FBLN2 | - | fibulin 2 | Reconstituted Complex | BioGRID | 11493006 |
| FGF2 | BFGF | FGFB | HBGF-2 | fibroblast growth factor 2 (basic) | - | HPRD | 11847221 |
| FGF7 | HBGF-7 | KGF | fibroblast growth factor 7 (keratinocyte growth factor) | - | HPRD,BioGRID | 10702276 |
| FGFBP1 | FGFBP | HBP17 | fibroblast growth factor binding protein 1 | - | HPRD,BioGRID | 11148217 |
| FN1 | CIG | DKFZp686F10164 | DKFZp686H0342 | DKFZp686I1370 | DKFZp686O13149 | ED-B | FINC | FN | FNZ | GFND | GFND2 | LETS | MSF | fibronectin 1 | Reconstituted Complex | BioGRID | 11493006 |
| ITGB1 | CD29 | FNRB | GPIIA | MDF2 | MSK12 | VLA-BETA | VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | - | HPRD | 9431988 |
| LAMA1 | LAMA | laminin, alpha 1 | - | HPRD,BioGRID | 9688542 |
| NID1 | NID | nidogen 1 | - | HPRD | 11493006 |
| NID2 | - | nidogen 2 (osteonidogen) | - | HPRD | 9733643 |
| NID2 | - | nidogen 2 (osteonidogen) | Reconstituted Complex | BioGRID | 11493006 |
| PDGFA | PDGF-A | PDGF1 | platelet-derived growth factor alpha polypeptide | in vitro | BioGRID | 9692901 |
| PDGFB | FLJ12858 | PDGF2 | SIS | SSV | c-sis | platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog) | - | HPRD,BioGRID | 9692901 |
| PRELP | MGC45323 | MST161 | MSTP161 | SLRR2A | proline/arginine-rich end leucine-rich repeat protein | - | HPRD,BioGRID | 11847210 |
| SPARC | ON | secreted protein, acidic, cysteine-rich (osteonectin) | - | HPRD | 2745554 |
| TTR | HsT2651 | PALB | TBPA | transthyretin | - | HPRD,BioGRID | 9307034 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ECM RECEPTOR INTERACTION | 84 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME HS GAG DEGRADATION | 20 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | 49 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME HS GAG BIOSYNTHESIS | 31 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | 52 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSAMINOGLYCAN METABOLISM | 111 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | 46 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPOPROTEIN METABOLISM | 28 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | 16 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME AMYLOIDS | 83 | 63 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| OXFORD RALA AND RALB TARGETS DN | 9 | 5 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA UP | 110 | 70 | All SZGR 2.0 genes in this pathway |
| LAMB CCND1 TARGETS | 19 | 14 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION UP | 71 | 51 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE DN | 64 | 45 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| MARIADASON REGULATED BY HISTONE ACETYLATION UP | 83 | 49 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT DN | 80 | 56 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C1 | 19 | 15 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA EARLY RESPONSE TO TGFB1 | 58 | 43 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| BONCI TARGETS OF MIR15A AND MIR16 1 | 91 | 75 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 13 | 30 | 20 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| AZARE NEOPLASTIC TRANSFORMATION BY STAT3 DN | 17 | 10 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| NABA PROTEOGLYCANS | 35 | 23 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA BASEMENT MEMBRANES | 40 | 22 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |