Gene Page: ACADM
Summary ?
| GeneID | 34 |
| Symbol | ACADM |
| Synonyms | ACAD1|MCAD|MCADH |
| Description | acyl-CoA dehydrogenase, C-4 to C-12 straight chain |
| Reference | MIM:607008|HGNC:HGNC:89|Ensembl:ENSG00000117054|HPRD:08447|Vega:OTTHUMG00000009784 |
| Gene type | protein-coding |
| Map location | 1p31 |
| Pascal p-value | 0.202 |
| Sherlock p-value | 0.048 |
| Fetal beta | 0.657 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.02692 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
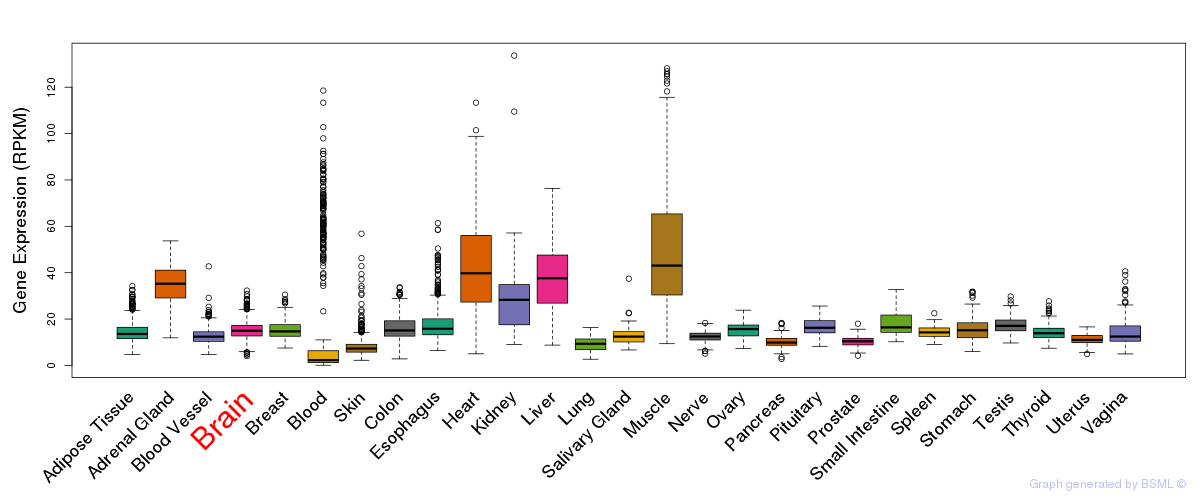
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MICALL2 | 0.79 | 0.83 |
| AMT | 0.77 | 0.79 |
| C1orf175 | 0.75 | 0.77 |
| MYO15B | 0.74 | 0.74 |
| GPT | 0.74 | 0.77 |
| RAPGEF3 | 0.74 | 0.80 |
| SLC27A1 | 0.73 | 0.71 |
| ITGA7 | 0.73 | 0.77 |
| AC024575.3 | 0.72 | 0.74 |
| ITIH4 | 0.72 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AZIN1 | -0.59 | -0.60 |
| HSPA13 | -0.58 | -0.58 |
| MOBKL3 | -0.58 | -0.59 |
| MAPK6 | -0.58 | -0.58 |
| ANKRD46 | -0.57 | -0.58 |
| VTA1 | -0.57 | -0.55 |
| IPMK | -0.57 | -0.55 |
| NUDT21 | -0.57 | -0.55 |
| AASDHPPT | -0.57 | -0.57 |
| DR1 | -0.57 | -0.58 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003995 | acyl-CoA dehydrogenase activity | IEA | - | |
| GO:0003995 | acyl-CoA dehydrogenase activity | IMP | 2393404 | |
| GO:0009055 | electron carrier activity | IEA | - | |
| GO:0050660 | FAD binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001889 | liver development | IEA | - | |
| GO:0055007 | cardiac muscle cell differentiation | IEA | - | |
| GO:0006082 | organic acid metabolic process | IEA | - | |
| GO:0009409 | response to cold | IEA | - | |
| GO:0009437 | carnitine metabolic process | IEA | - | |
| GO:0009791 | post-embryonic development | IEA | - | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0007507 | heart development | IEA | - | |
| GO:0006635 | fatty acid beta-oxidation | IMP | 2393404 | |
| GO:0006631 | fatty acid metabolic process | IEA | - | |
| GO:0006629 | lipid metabolic process | IEA | - | |
| GO:0042594 | response to starvation | IEA | - | |
| GO:0055114 | oxidation reduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005739 | mitochondrion | TAS | 1731887 | |
| GO:0005759 | mitochondrial matrix | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FATTY ACID METABOLISM | 42 | 29 | All SZGR 2.0 genes in this pathway |
| KEGG VALINE LEUCINE AND ISOLEUCINE DEGRADATION | 44 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG BETA ALANINE METABOLISM | 22 | 16 | All SZGR 2.0 genes in this pathway |
| KEGG PROPANOATE METABOLISM | 33 | 22 | All SZGR 2.0 genes in this pathway |
| KEGG PPAR SIGNALING PATHWAY | 69 | 47 | All SZGR 2.0 genes in this pathway |
| PID HNF3B PATHWAY | 45 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| CASTELLANO NRAS TARGETS UP | 68 | 41 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| OUELLET CULTURED OVARIAN CANCER INVASIVE VS LMP UP | 69 | 40 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 5 | 13 | 5 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER SMALL VS HUGE UP | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| GOLUB ALL VS AML UP | 24 | 20 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF DN | 84 | 50 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 5 | 126 | 78 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC UP | 123 | 75 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| GENTILE RESPONSE CLUSTER D3 | 61 | 39 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| SCHRAETS MLL TARGETS DN | 33 | 24 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MOOTHA FFA OXYDATION | 22 | 13 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 36HR | 152 | 88 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND WITH H4K20ME1 MARK | 145 | 82 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP | 149 | 96 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| SANDERSON PPARA TARGETS | 15 | 7 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |