Gene Page: IGF1
Summary ?
| GeneID | 3479 |
| Symbol | IGF1 |
| Synonyms | IGF-I|IGFI|MGF |
| Description | insulin like growth factor 1 |
| Reference | MIM:147440|HGNC:HGNC:5464|Ensembl:ENSG00000017427|HPRD:00936|Vega:OTTHUMG00000149910 |
| Gene type | protein-coding |
| Map location | 12q23.2 |
| Pascal p-value | 0.339 |
| Fetal beta | -1.233 |
| eGene | Cerebellar Hemisphere Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1911875 | chr4 | 132293356 | IGF1 | 3479 | 0.12 | trans |
Section II. Transcriptome annotation
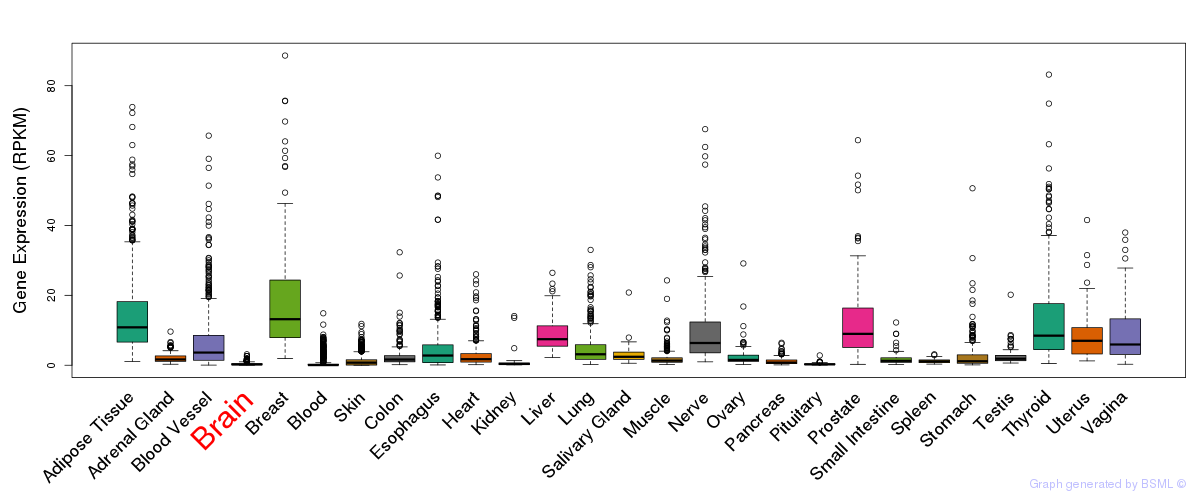
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005179 | hormone activity | IEA | - | |
| GO:0005179 | hormone activity | TAS | 9681507 | |
| GO:0005159 | insulin-like growth factor receptor binding | IPI | 8452530 | |
| GO:0005159 | insulin-like growth factor receptor binding | TAS | 10749889 | |
| GO:0005158 | insulin receptor binding | IPI | 8452530 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 10766744 | |
| GO:0008083 | growth factor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0010001 | glial cell differentiation | IEA | Glial (GO term level: 8) | - |
| GO:0007399 | nervous system development | IEA | neurite (GO term level: 5) | - |
| GO:0001501 | skeletal system development | TAS | 10448861 | |
| GO:0030104 | water homeostasis | IEA | - | |
| GO:0007265 | Ras protein signal transduction | TAS | 10848592 | |
| GO:0006260 | DNA replication | TAS | 10766744 | |
| GO:0009887 | organ morphogenesis | IEA | - | |
| GO:0007165 | signal transduction | TAS | 10448861 | |
| GO:0048468 | cell development | IEA | - | |
| GO:0007517 | muscle development | TAS | 10448861 | |
| GO:0007605 | sensory perception of sound | IEA | - | |
| GO:0009441 | glycolate metabolic process | TAS | 10448861 | |
| GO:0048015 | phosphoinositide-mediated signaling | IDA | 7692086 | |
| GO:0006928 | cell motion | TAS | 10766744 | |
| GO:0006916 | anti-apoptosis | IDA | 16942485 | |
| GO:0006916 | anti-apoptosis | IEA | - | |
| GO:0014065 | phosphoinositide 3-kinase cascade | IDA | 7688386 | |
| GO:0042523 | positive regulation of tyrosine phosphorylation of Stat5 protein | IDA | 9722506 | |
| GO:0033143 | regulation of steroid hormone receptor signaling pathway | IDA | 10459853 | |
| GO:0014834 | satellite cell maintenance involved in skeletal muscle regeneration | IDA | 17531227 | |
| GO:0014896 | muscle hypertrophy | IMP | 10191278 | |
| GO:0014904 | myotube cell development | IDA | 17531227 | |
| GO:0014911 | positive regulation of smooth muscle cell migration | IDA | 10766744 | |
| GO:0021940 | positive regulation of granule cell precursor proliferation | IEA | - | |
| GO:0043193 | positive regulation of gene-specific transcription | IDA | 9722506 | |
| GO:0048754 | branching morphogenesis of a tube | IEA | - | |
| GO:0051246 | regulation of protein metabolic process | IEA | - | |
| GO:0030879 | mammary gland development | IEA | - | |
| GO:0034392 | negative regulation of smooth muscle cell apoptosis | IDA | 16942485 | |
| GO:0035264 | multicellular organism growth | IEA | - | |
| GO:0045740 | positive regulation of DNA replication | IDA | 7688386 |9722506 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0045840 | positive regulation of mitosis | IDA | 7188854 | |
| GO:0048146 | positive regulation of fibroblast proliferation | IDA | 7188854 | |
| GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process | IEA | - | |
| GO:0050679 | positive regulation of epithelial cell proliferation | IDA | 7188854 | |
| GO:0045821 | positive regulation of glycolysis | IDA | 7688386 | |
| GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway | IDA | 7688386 |10766744 | |
| GO:0046579 | positive regulation of Ras protein signal transduction | IDA | 9722506 | |
| GO:0048661 | positive regulation of smooth muscle cell proliferation | IDA | 10766744 |16942485 | |
| GO:0045445 | myoblast differentiation | IDA | 17531227 | |
| GO:0051450 | myoblast proliferation | IDA | 17531227 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | EXP | 2752154 | |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005576 | extracellular region | NAS | 14718574 | |
| GO:0005615 | extracellular space | IEA | - | |
| GO:0016942 | insulin-like growth factor binding protein complex | IC | 10766744 | |
| GO:0031093 | platelet alpha granule lumen | EXP | 2752154 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| IDE | FLJ35968 | INSULYSIN | insulin-degrading enzyme | Reconstituted Complex | BioGRID | 1733942 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | IGF-I interacts with IGF-IR. This interaction was modeled on a demonstrated interaction between IGF-I from an unspecified species and human IGF-IR. | BIND | 15604363 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | - | HPRD,BioGRID | 1852007 |
| IGFALS | ALS | insulin-like growth factor binding protein, acid labile subunit | - | HPRD,BioGRID | 9497324 |10823924 |
| IGFBP1 | AFBP | IBP1 | IGF-BP25 | PP12 | hIGFBP-1 | insulin-like growth factor binding protein 1 | - | HPRD,BioGRID | 10350456 |
| IGFBP1 | AFBP | IBP1 | IGF-BP25 | PP12 | hIGFBP-1 | insulin-like growth factor binding protein 1 | IGF-I interacts with IGFBP-1. | BIND | 10407151 |
| IGFBP2 | IBP2 | IGF-BP53 | insulin-like growth factor binding protein 2, 36kDa | IGF-I interacts with IGFBP-2. | BIND | 10407151 |
| IGFBP2 | IBP2 | IGF-BP53 | insulin-like growth factor binding protein 2, 36kDa | - | HPRD,BioGRID | 11063745 |
| IGFBP3 | BP-53 | IBP3 | insulin-like growth factor binding protein 3 | - | HPRD,BioGRID | 11600567 |
| IGFBP3 | BP-53 | IBP3 | insulin-like growth factor binding protein 3 | IGF-I interacts with IGFBP-3. | BIND | 10407151 |
| IGFBP4 | BP-4 | HT29-IGFBP | IBP4 | IGFBP-4 | insulin-like growth factor binding protein 4 | - | HPRD,BioGRID | 9722589 |
| IGFBP4 | BP-4 | HT29-IGFBP | IBP4 | IGFBP-4 | insulin-like growth factor binding protein 4 | IGF-I interacts with IGFBP-4. | BIND | 10407151 |
| IGFBP5 | IBP5 | insulin-like growth factor binding protein 5 | IGF-I interacts with IGFBP-5. | BIND | 10407151 |
| IGFBP5 | IBP5 | insulin-like growth factor binding protein 5 | - | HPRD,BioGRID | 9497324 |
| IGFBP6 | IBP6 | insulin-like growth factor binding protein 6 | IGF-I interacts with IGFBP-6. | BIND | 10407151 |
| IGFBP6 | IBP6 | insulin-like growth factor binding protein 6 | - | HPRD | 7683646 |
| IGFBP7 | FSTL2 | IGFBP-7 | IGFBP-7v | IGFBPRP1 | MAC25 | PSF | insulin-like growth factor binding protein 7 | - | HPRD,BioGRID | 8939990 |
| IGSF1 | IGCD1 | IGDC1 | INHBP | KIAA0364 | MGC75490 | PGSF2 | immunoglobulin superfamily, member 1 | - | HPRD,BioGRID | 11344214 |
| NOV | CCN3 | IGFBP9 | nephroblastoma overexpressed gene | - | HPRD,BioGRID | 10084601 |
| TF | DKFZp781D0156 | PRO1557 | PRO2086 | transferrin | Reconstituted Complex | BioGRID | 11749962 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG MTOR SIGNALING PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG PROGESTERONE MEDIATED OOCYTE MATURATION | 86 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG ALDOSTERONE REGULATED SODIUM REABSORPTION | 42 | 29 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HDAC PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERYTH PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1 PATHWAY | 21 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFAT PATHWAY | 56 | 45 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BAD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1MTOR PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA LONGEVITY PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| PID ER NONGENOMIC PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| PID SHP2 PATHWAY | 58 | 46 | All SZGR 2.0 genes in this pathway |
| PID IGF1 PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID CERAMIDE PATHWAY | 48 | 37 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL UP | 69 | 38 | All SZGR 2.0 genes in this pathway |
| CORRE MULTIPLE MYELOMA DN | 62 | 41 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| PACHER TARGETS OF IGF1 AND IGF2 UP | 35 | 27 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| BARRIER CANCER RELAPSE NORMAL SAMPLE UP | 32 | 21 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| BUSA SAM68 TARGETS DN | 6 | 5 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 UP | 30 | 24 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN 2FC DN | 21 | 13 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| GALIE TUMOR ANGIOGENESIS | 8 | 7 | All SZGR 2.0 genes in this pathway |
| BERTUCCI INVASIVE CARCINOMA DUCTAL VS LOBULAR DN | 46 | 34 | All SZGR 2.0 genes in this pathway |
| GARCIA TARGETS OF FLI1 AND DAX1 DN | 176 | 104 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER SOX9 TARGETS IN PROSTATE DEVELOPMENT DN | 45 | 33 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER F | 54 | 32 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA DN | 65 | 44 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 DN | 64 | 38 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC DN | 63 | 40 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 DN | 64 | 42 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| GUO HEX TARGETS DN | 65 | 36 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS DN | 16 | 11 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| LIU SMARCA4 TARGETS | 64 | 39 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 0HR | 63 | 48 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP | 113 | 71 | All SZGR 2.0 genes in this pathway |
| BACOLOD RESISTANCE TO ALKYLATING AGENTS UP | 26 | 17 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS UP | 49 | 40 | All SZGR 2.0 genes in this pathway |
| WENG POR TARGETS LIVER DN | 20 | 11 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR UV | 63 | 44 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN LUNG DN | 37 | 22 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL A UP | 84 | 52 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| MATZUK EARLY ANTRAL FOLLICLE | 13 | 11 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH DN | 31 | 25 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| WU SILENCED BY METHYLATION IN BLADDER CANCER | 55 | 42 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| ZHAN LATE DIFFERENTIATION GENES UP | 33 | 24 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL DN | 113 | 76 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 ICP WITH H3K4ME3 AND H3K27ME3 | 34 | 21 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA FGF2 TARGETS | 29 | 17 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 DN | 80 | 51 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 AND H3K27ME3 | 137 | 85 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 11 | 103 | 68 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| TERAO AOX4 TARGETS SKIN DN | 27 | 15 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR1 TARGETS UP | 53 | 39 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 151 | 158 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-142-5p | 4421 | 4428 | 1A,m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-150 | 3590 | 3597 | 1A,m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-18 | 181 | 187 | m8 | hsa-miR-18a | UAAGGUGCAUCUAGUGCAGAUA |
| hsa-miR-18b | UAAGGUGCAUCUAGUGCAGUUA | ||||
| miR-186 | 3676 | 3682 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU | ||||
| miR-19 | 178 | 184 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-26 | 3689 | 3695 | m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-27 | 4376 | 4382 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-29 | 3664 | 3671 | 1A,m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU | ||||
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-30-3p | 3278 | 3284 | 1A | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-320 | 640 | 646 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-409-3p | 150 | 156 | m8 | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-486 | 855 | 861 | 1A | hsa-miR-486 | UCCUGUACUGAGCUGCCCCGAG |
| miR-495 | 916 | 923 | 1A,m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-496 | 832 | 838 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| hsa-miR-496 | AUUACAUGGCCAAUCUC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.